User login
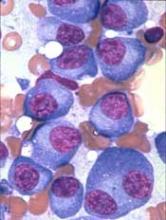
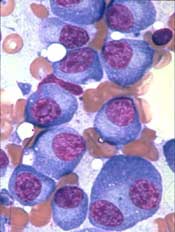
Researchers have identified novel genes involved in the development of multiple myeloma (MM), according to a paper published in Leukemia.
The team’s analyses revealed regions of coding and non-coding DNA that appear to drive MM development.
The researchers analyzed whole-exome sequencing data from 804 MM patients and whole-genome sequencing data from 765 MM patients.
This revealed 16 novel genes that were disrupted in coding regions of DNA and 15 novel genes disrupted in non-coding regions.
There were 5 genes disrupted by structural variants in coding regions—CD96, PRDM1, FBXW7, MAP3K14, and CCND2.
There were also 11 genes disrupted by single nucleotide variants (SNVs) and indels in coding regions—BAX, C8orf86, FAM154B, FTL, HIST1H4H, LEMD2, PABPC1, RPN1, RPS3A, SGPP1, and TBC1D29.
Among the novel genes disrupted by mutations in non-coding regions was NBPF1, a promoter disrupted by SNVs. The researchers noted that NBPF1 is directly regulated by NF-κB, and the NF-κB pathway is recurrently affected in MM.
The team also identified 7 cis-regulatory elements (CREs) disrupted by SNVs—CALCB, COBLL1, HOXB3, ST6GAL1, PAX5, ATP13A2, and TPRG1.
The researchers said the SNVs in PAX5 and HOXB3 reduced gene expression, suggesting PAX5 and HOXB3 function as tumor suppressors in MM. On the other hand, the SNVs in ST6GAL1 increased gene expression, which may contribute to the aberrant immunoglobulin-G glycosylation seen in MM.
Finally, there were 7 CREs disrupted by copy number variations—MYC, PLD4, KDM3B, SP110, RAB36, PACS2, and TEX22.
The researchers noted that, with the exception of MYC, these genes reside close to regions of common structural variation, so their relevance in MM is not clear.
The team also said it’s well known that MYC is upregulated in MM through gene amplification or translocation, but this research shows that MYC can be dysregulated by alternative mechanisms.
“We need smarter, kinder treatments for myeloma that are more tailored to each person’s cancer,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“Exhaustive genetic research like this is helping us to make that possible. Our findings should now open up new avenues for discovering treatments that target the genes driving myeloma.”
Researchers have identified novel genes involved in the development of multiple myeloma (MM), according to a paper published in Leukemia.
The team’s analyses revealed regions of coding and non-coding DNA that appear to drive MM development.
The researchers analyzed whole-exome sequencing data from 804 MM patients and whole-genome sequencing data from 765 MM patients.
This revealed 16 novel genes that were disrupted in coding regions of DNA and 15 novel genes disrupted in non-coding regions.
There were 5 genes disrupted by structural variants in coding regions—CD96, PRDM1, FBXW7, MAP3K14, and CCND2.
There were also 11 genes disrupted by single nucleotide variants (SNVs) and indels in coding regions—BAX, C8orf86, FAM154B, FTL, HIST1H4H, LEMD2, PABPC1, RPN1, RPS3A, SGPP1, and TBC1D29.
Among the novel genes disrupted by mutations in non-coding regions was NBPF1, a promoter disrupted by SNVs. The researchers noted that NBPF1 is directly regulated by NF-κB, and the NF-κB pathway is recurrently affected in MM.
The team also identified 7 cis-regulatory elements (CREs) disrupted by SNVs—CALCB, COBLL1, HOXB3, ST6GAL1, PAX5, ATP13A2, and TPRG1.
The researchers said the SNVs in PAX5 and HOXB3 reduced gene expression, suggesting PAX5 and HOXB3 function as tumor suppressors in MM. On the other hand, the SNVs in ST6GAL1 increased gene expression, which may contribute to the aberrant immunoglobulin-G glycosylation seen in MM.
Finally, there were 7 CREs disrupted by copy number variations—MYC, PLD4, KDM3B, SP110, RAB36, PACS2, and TEX22.
The researchers noted that, with the exception of MYC, these genes reside close to regions of common structural variation, so their relevance in MM is not clear.
The team also said it’s well known that MYC is upregulated in MM through gene amplification or translocation, but this research shows that MYC can be dysregulated by alternative mechanisms.
“We need smarter, kinder treatments for myeloma that are more tailored to each person’s cancer,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“Exhaustive genetic research like this is helping us to make that possible. Our findings should now open up new avenues for discovering treatments that target the genes driving myeloma.”
Researchers have identified novel genes involved in the development of multiple myeloma (MM), according to a paper published in Leukemia.
The team’s analyses revealed regions of coding and non-coding DNA that appear to drive MM development.
The researchers analyzed whole-exome sequencing data from 804 MM patients and whole-genome sequencing data from 765 MM patients.
This revealed 16 novel genes that were disrupted in coding regions of DNA and 15 novel genes disrupted in non-coding regions.
There were 5 genes disrupted by structural variants in coding regions—CD96, PRDM1, FBXW7, MAP3K14, and CCND2.
There were also 11 genes disrupted by single nucleotide variants (SNVs) and indels in coding regions—BAX, C8orf86, FAM154B, FTL, HIST1H4H, LEMD2, PABPC1, RPN1, RPS3A, SGPP1, and TBC1D29.
Among the novel genes disrupted by mutations in non-coding regions was NBPF1, a promoter disrupted by SNVs. The researchers noted that NBPF1 is directly regulated by NF-κB, and the NF-κB pathway is recurrently affected in MM.
The team also identified 7 cis-regulatory elements (CREs) disrupted by SNVs—CALCB, COBLL1, HOXB3, ST6GAL1, PAX5, ATP13A2, and TPRG1.
The researchers said the SNVs in PAX5 and HOXB3 reduced gene expression, suggesting PAX5 and HOXB3 function as tumor suppressors in MM. On the other hand, the SNVs in ST6GAL1 increased gene expression, which may contribute to the aberrant immunoglobulin-G glycosylation seen in MM.
Finally, there were 7 CREs disrupted by copy number variations—MYC, PLD4, KDM3B, SP110, RAB36, PACS2, and TEX22.
The researchers noted that, with the exception of MYC, these genes reside close to regions of common structural variation, so their relevance in MM is not clear.
The team also said it’s well known that MYC is upregulated in MM through gene amplification or translocation, but this research shows that MYC can be dysregulated by alternative mechanisms.
“We need smarter, kinder treatments for myeloma that are more tailored to each person’s cancer,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“Exhaustive genetic research like this is helping us to make that possible. Our findings should now open up new avenues for discovering treatments that target the genes driving myeloma.”