User login
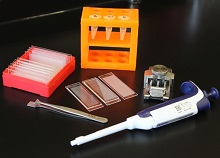
portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()

portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()

portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()