User login
A new mathematical model could aid the development of drugs that target malaria parasites’ metabolic processes, according to researchers.
The team said they developed the first model of a malaria parasite that accurately integrates its genetics and metabolism.
They described this model in PLOS Computational Biology.
“The model integrates all available knowledge on the genetics and metabolism of the parasites and allows the formulation of testable hypotheses behind the parasites’ essential functions,” said study author Vassily Hatzimanikatis, PhD, of École Polytechnique Fédérale de Lausanne in Switzerland.
“Ultimately, it can accelerate the discovery toward novel antimalarial drug targets.”
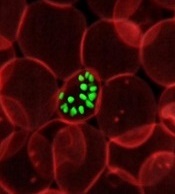
To create the model, Dr Hatzimanikatis and his colleagues studied Plasmodium falciparum parasites, focusing on the way they produce and use energy for their metabolic reactions.
This revealed which metabolic functions were essential at each stage of infection and which were energetically coupled through key metabolites.
The team was therefore able to model the bioenergetics of the metabolism of P falciparum, predicting which genes are indispensable for every biological function in the parasite.
By integrating metabolomics and genetics data, the model revealed the complex interactions between gene products, reactions, and metabolites in the parasite, and it revealed potential mechanisms to target with drugs.
“The design of efficient antimalarial drugs that target the parasite’s and not the patient’s metabolism requires an in-depth understanding of the mechanisms that make a particular enzyme essential,” said Anush Chiappino-Pepe, a PhD student at École Polytechnique Fédérale de Lausanne.
“So mathematical modeling of the parasite’s metabolism becomes a very powerful tool.”
The researchers said they will continue to calibrate and improve the predictive capabilities of their model with additional genetics and metabolomics data.
They hope to reveal the mechanisms behind host-pathogen interactions and gain insight into the physiology of P falciparum while it is dormant. ![]()
A new mathematical model could aid the development of drugs that target malaria parasites’ metabolic processes, according to researchers.
The team said they developed the first model of a malaria parasite that accurately integrates its genetics and metabolism.
They described this model in PLOS Computational Biology.
“The model integrates all available knowledge on the genetics and metabolism of the parasites and allows the formulation of testable hypotheses behind the parasites’ essential functions,” said study author Vassily Hatzimanikatis, PhD, of École Polytechnique Fédérale de Lausanne in Switzerland.
“Ultimately, it can accelerate the discovery toward novel antimalarial drug targets.”
To create the model, Dr Hatzimanikatis and his colleagues studied Plasmodium falciparum parasites, focusing on the way they produce and use energy for their metabolic reactions.
This revealed which metabolic functions were essential at each stage of infection and which were energetically coupled through key metabolites.
The team was therefore able to model the bioenergetics of the metabolism of P falciparum, predicting which genes are indispensable for every biological function in the parasite.
By integrating metabolomics and genetics data, the model revealed the complex interactions between gene products, reactions, and metabolites in the parasite, and it revealed potential mechanisms to target with drugs.
“The design of efficient antimalarial drugs that target the parasite’s and not the patient’s metabolism requires an in-depth understanding of the mechanisms that make a particular enzyme essential,” said Anush Chiappino-Pepe, a PhD student at École Polytechnique Fédérale de Lausanne.
“So mathematical modeling of the parasite’s metabolism becomes a very powerful tool.”
The researchers said they will continue to calibrate and improve the predictive capabilities of their model with additional genetics and metabolomics data.
They hope to reveal the mechanisms behind host-pathogen interactions and gain insight into the physiology of P falciparum while it is dormant. ![]()
A new mathematical model could aid the development of drugs that target malaria parasites’ metabolic processes, according to researchers.
The team said they developed the first model of a malaria parasite that accurately integrates its genetics and metabolism.
They described this model in PLOS Computational Biology.
“The model integrates all available knowledge on the genetics and metabolism of the parasites and allows the formulation of testable hypotheses behind the parasites’ essential functions,” said study author Vassily Hatzimanikatis, PhD, of École Polytechnique Fédérale de Lausanne in Switzerland.
“Ultimately, it can accelerate the discovery toward novel antimalarial drug targets.”
To create the model, Dr Hatzimanikatis and his colleagues studied Plasmodium falciparum parasites, focusing on the way they produce and use energy for their metabolic reactions.
This revealed which metabolic functions were essential at each stage of infection and which were energetically coupled through key metabolites.
The team was therefore able to model the bioenergetics of the metabolism of P falciparum, predicting which genes are indispensable for every biological function in the parasite.
By integrating metabolomics and genetics data, the model revealed the complex interactions between gene products, reactions, and metabolites in the parasite, and it revealed potential mechanisms to target with drugs.
“The design of efficient antimalarial drugs that target the parasite’s and not the patient’s metabolism requires an in-depth understanding of the mechanisms that make a particular enzyme essential,” said Anush Chiappino-Pepe, a PhD student at École Polytechnique Fédérale de Lausanne.
“So mathematical modeling of the parasite’s metabolism becomes a very powerful tool.”
The researchers said they will continue to calibrate and improve the predictive capabilities of their model with additional genetics and metabolomics data.
They hope to reveal the mechanisms behind host-pathogen interactions and gain insight into the physiology of P falciparum while it is dormant. ![]()