User login
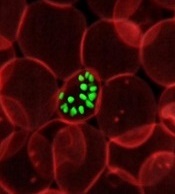
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()