User login
Some children are more susceptible to viral and bacterial respiratory infections in the first few years of life than others. However, the factors contributing to this susceptibility are incompletely understood. The pathogenesis, development, severity, and clinical outcomes of respiratory infections are largely dependent on the resident composition of the nasopharyngeal microbiome and immune defense.1
Respiratory infections caused by bacteria and/or viruses are a leading cause of death in children in the United States and worldwide. The well-recognized, predominant causative bacteria are Streptococcus pneumoniae (pneumococcus), nontypeable Haemophilus influenzae (Hflu), and Moraxella catarrhalis (Mcat). Respiratory infections caused by these pathogens result in considerable morbidity, mortality, and account for high health care costs. The clinical and laboratory group that I lead in Rochester, N.Y., has been studying acute otitis media (AOM) etiology, epidemiology, pathogenesis, prevention, and treatment for over 3 decades. Our research findings are likely applicable and generalizable to understanding the pathogenesis and immune response to other infectious diseases induced by pneumococcus, Hflu, and Mcat since they are also key pathogens causing sinusitis and lung infections.
Previous immunologic analysis of children with AOM by our group provided clarity in differences between infection-prone children manifest as otitis prone (OP; often referred to in our publications as stringently defined OP because of the stringent diagnostic requirement of tympanocentesis-proven etiology of infection) and non-OP children. We showed that about 90% of OP children have deficient immune responses following nasopharyngeal colonization and AOM, demonstrated by inadequate innate responses and adaptive immune responses.2 Many of these children also showed an increased propensity to viral upper respiratory infection and 30% fail to produce protective antibody responses after injection of routine pediatric vaccines.3,4
In this column, I want to share new information regarding differences in the nasopharyngeal microbiome of children who are respiratory infection prone versus those who are non–respiratory infection prone and children with asthma versus those who do not exhibit that clinical phenotype. We performed a retrospective analysis of clinical samples collected from 358 children, aged 6 months to 5 years, from our prospectively enrolled cohort in Rochester, N.Y., to determine associations between AOM and other childhood respiratory illnesses and nasopharyngeal microbiota. In order to define subgroups of children within the cohort, we used a statistical method called unsupervised clustering analysis to see if relatively unique groups of children could be discerned. The overall cohort successfully clustered into two groups, showing marked differences in the prevalence of respiratory infections and asthma.5 We termed the two clinical phenotypes infection and asthma prone (n = 99, 28% of the children) and non–infection and asthma prone (n = 259, 72% of the children). Infection- and asthma-prone children were significantly more likely to experience recurrent AOM, influenza, sinusitis, pneumonia, asthma, and allergic rhinitis, compared with non–infection- and asthma-prone children (Figure).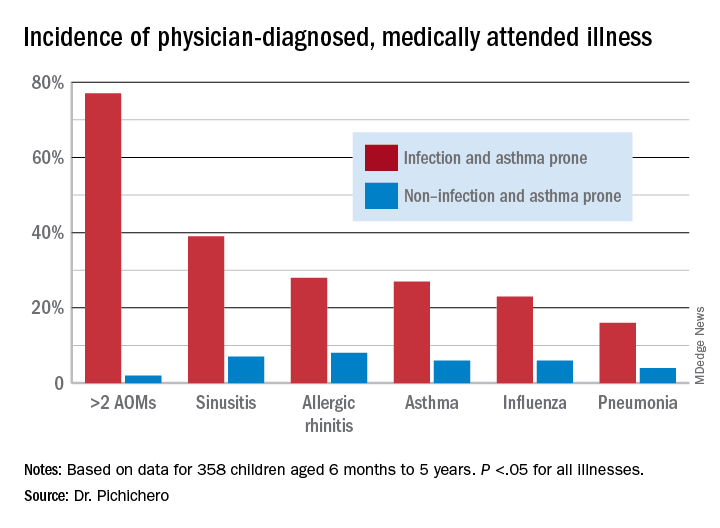
The two groups did not experience significantly different rates of eczema, food allergy, skin infections, urinary tract infections, or acute gastroenteritis, suggesting a common thread involving the respiratory tract that did not cross over to the gastrointestinal, skin, or urinary tract. We found that age at first nasopharyngeal colonization with any of the three bacterial respiratory pathogens (pneumococcus, Hflu, or Mcat) was significantly associated with the respiratory infection– and asthma-prone clinical phenotype. Specifically, respiratory infection– and asthma-prone children experienced colonization at a significantly earlier age than nonprone children did for all three bacteria. In an analysis of individual conditions, early Mcat colonization significantly associated with pneumonia, sinusitis, and asthma susceptibility; Hflu with pneumonia, sinusitis, influenza, and allergic rhinitis; and pneumococcus with sinusitis.
Since early colonization with the three bacterial respiratory pathogens was strongly associated with respiratory illnesses and asthma, nasopharyngeal microbiome analysis was performed on an available subset of samples. Bacterial diversity trended lower in infection- and asthma-prone children, consistent with dysbiosis in the respiratory infection– and asthma-prone clinical phenotype. Nine different bacteria genera were found to be differentially abundant when comparing respiratory infection– and asthma-prone and nonprone children, pointing the way to possible interventions to make the respiratory infection– and asthma-prone child nasopharyngeal microbiome more like the nonprone child.
As I have written previously in this column, recent accumulating data have shed light on the importance of the human microbiome in modulating immune homeostasis and disease susceptibility.6 My group is working toward generating new knowledge for the long-term goal of identifying new therapeutic strategies to facilitate a protective, diverse nasopharyngeal microbiome (with appropriately tuned intranasal probiotics) to prevent respiratory pathogen colonization and/or subsequent progression to respiratory infection and asthma. Also, vaccines directed against colonization-enhancing members of the microbiome may provide a means to indirectly control respiratory pathogen nasopharyngeal colonization.
Dr. Pichichero is a specialist in pediatric infectious diseases and director of the Research Institute at Rochester (N.Y.) General Hospital. He has no conflicts to declare. Contact him at [email protected]
References
1. Man WH et al. Nat Rev Microbiol. 2017;15(5):259-70.
2. Pichichero ME. J Infect. 2020;80(6):614-22.
3. Ren D et al. Clin Infect Dis. 2019;68(9):1566-74.
4. Pichichero ME et al. Pediatr Infect Dis J. 2013;32(11):1163-8.
5. Chapman T et al. PLoS One. 2020 Dec 11;15(12).
6. Blaser MJ. The microbiome revolution. J Clin Invest. 2014;124:4162-5.
Some children are more susceptible to viral and bacterial respiratory infections in the first few years of life than others. However, the factors contributing to this susceptibility are incompletely understood. The pathogenesis, development, severity, and clinical outcomes of respiratory infections are largely dependent on the resident composition of the nasopharyngeal microbiome and immune defense.1
Respiratory infections caused by bacteria and/or viruses are a leading cause of death in children in the United States and worldwide. The well-recognized, predominant causative bacteria are Streptococcus pneumoniae (pneumococcus), nontypeable Haemophilus influenzae (Hflu), and Moraxella catarrhalis (Mcat). Respiratory infections caused by these pathogens result in considerable morbidity, mortality, and account for high health care costs. The clinical and laboratory group that I lead in Rochester, N.Y., has been studying acute otitis media (AOM) etiology, epidemiology, pathogenesis, prevention, and treatment for over 3 decades. Our research findings are likely applicable and generalizable to understanding the pathogenesis and immune response to other infectious diseases induced by pneumococcus, Hflu, and Mcat since they are also key pathogens causing sinusitis and lung infections.
Previous immunologic analysis of children with AOM by our group provided clarity in differences between infection-prone children manifest as otitis prone (OP; often referred to in our publications as stringently defined OP because of the stringent diagnostic requirement of tympanocentesis-proven etiology of infection) and non-OP children. We showed that about 90% of OP children have deficient immune responses following nasopharyngeal colonization and AOM, demonstrated by inadequate innate responses and adaptive immune responses.2 Many of these children also showed an increased propensity to viral upper respiratory infection and 30% fail to produce protective antibody responses after injection of routine pediatric vaccines.3,4
In this column, I want to share new information regarding differences in the nasopharyngeal microbiome of children who are respiratory infection prone versus those who are non–respiratory infection prone and children with asthma versus those who do not exhibit that clinical phenotype. We performed a retrospective analysis of clinical samples collected from 358 children, aged 6 months to 5 years, from our prospectively enrolled cohort in Rochester, N.Y., to determine associations between AOM and other childhood respiratory illnesses and nasopharyngeal microbiota. In order to define subgroups of children within the cohort, we used a statistical method called unsupervised clustering analysis to see if relatively unique groups of children could be discerned. The overall cohort successfully clustered into two groups, showing marked differences in the prevalence of respiratory infections and asthma.5 We termed the two clinical phenotypes infection and asthma prone (n = 99, 28% of the children) and non–infection and asthma prone (n = 259, 72% of the children). Infection- and asthma-prone children were significantly more likely to experience recurrent AOM, influenza, sinusitis, pneumonia, asthma, and allergic rhinitis, compared with non–infection- and asthma-prone children (Figure).
The two groups did not experience significantly different rates of eczema, food allergy, skin infections, urinary tract infections, or acute gastroenteritis, suggesting a common thread involving the respiratory tract that did not cross over to the gastrointestinal, skin, or urinary tract. We found that age at first nasopharyngeal colonization with any of the three bacterial respiratory pathogens (pneumococcus, Hflu, or Mcat) was significantly associated with the respiratory infection– and asthma-prone clinical phenotype. Specifically, respiratory infection– and asthma-prone children experienced colonization at a significantly earlier age than nonprone children did for all three bacteria. In an analysis of individual conditions, early Mcat colonization significantly associated with pneumonia, sinusitis, and asthma susceptibility; Hflu with pneumonia, sinusitis, influenza, and allergic rhinitis; and pneumococcus with sinusitis.
Since early colonization with the three bacterial respiratory pathogens was strongly associated with respiratory illnesses and asthma, nasopharyngeal microbiome analysis was performed on an available subset of samples. Bacterial diversity trended lower in infection- and asthma-prone children, consistent with dysbiosis in the respiratory infection– and asthma-prone clinical phenotype. Nine different bacteria genera were found to be differentially abundant when comparing respiratory infection– and asthma-prone and nonprone children, pointing the way to possible interventions to make the respiratory infection– and asthma-prone child nasopharyngeal microbiome more like the nonprone child.
As I have written previously in this column, recent accumulating data have shed light on the importance of the human microbiome in modulating immune homeostasis and disease susceptibility.6 My group is working toward generating new knowledge for the long-term goal of identifying new therapeutic strategies to facilitate a protective, diverse nasopharyngeal microbiome (with appropriately tuned intranasal probiotics) to prevent respiratory pathogen colonization and/or subsequent progression to respiratory infection and asthma. Also, vaccines directed against colonization-enhancing members of the microbiome may provide a means to indirectly control respiratory pathogen nasopharyngeal colonization.
Dr. Pichichero is a specialist in pediatric infectious diseases and director of the Research Institute at Rochester (N.Y.) General Hospital. He has no conflicts to declare. Contact him at [email protected]
References
1. Man WH et al. Nat Rev Microbiol. 2017;15(5):259-70.
2. Pichichero ME. J Infect. 2020;80(6):614-22.
3. Ren D et al. Clin Infect Dis. 2019;68(9):1566-74.
4. Pichichero ME et al. Pediatr Infect Dis J. 2013;32(11):1163-8.
5. Chapman T et al. PLoS One. 2020 Dec 11;15(12).
6. Blaser MJ. The microbiome revolution. J Clin Invest. 2014;124:4162-5.
Some children are more susceptible to viral and bacterial respiratory infections in the first few years of life than others. However, the factors contributing to this susceptibility are incompletely understood. The pathogenesis, development, severity, and clinical outcomes of respiratory infections are largely dependent on the resident composition of the nasopharyngeal microbiome and immune defense.1
Respiratory infections caused by bacteria and/or viruses are a leading cause of death in children in the United States and worldwide. The well-recognized, predominant causative bacteria are Streptococcus pneumoniae (pneumococcus), nontypeable Haemophilus influenzae (Hflu), and Moraxella catarrhalis (Mcat). Respiratory infections caused by these pathogens result in considerable morbidity, mortality, and account for high health care costs. The clinical and laboratory group that I lead in Rochester, N.Y., has been studying acute otitis media (AOM) etiology, epidemiology, pathogenesis, prevention, and treatment for over 3 decades. Our research findings are likely applicable and generalizable to understanding the pathogenesis and immune response to other infectious diseases induced by pneumococcus, Hflu, and Mcat since they are also key pathogens causing sinusitis and lung infections.
Previous immunologic analysis of children with AOM by our group provided clarity in differences between infection-prone children manifest as otitis prone (OP; often referred to in our publications as stringently defined OP because of the stringent diagnostic requirement of tympanocentesis-proven etiology of infection) and non-OP children. We showed that about 90% of OP children have deficient immune responses following nasopharyngeal colonization and AOM, demonstrated by inadequate innate responses and adaptive immune responses.2 Many of these children also showed an increased propensity to viral upper respiratory infection and 30% fail to produce protective antibody responses after injection of routine pediatric vaccines.3,4
In this column, I want to share new information regarding differences in the nasopharyngeal microbiome of children who are respiratory infection prone versus those who are non–respiratory infection prone and children with asthma versus those who do not exhibit that clinical phenotype. We performed a retrospective analysis of clinical samples collected from 358 children, aged 6 months to 5 years, from our prospectively enrolled cohort in Rochester, N.Y., to determine associations between AOM and other childhood respiratory illnesses and nasopharyngeal microbiota. In order to define subgroups of children within the cohort, we used a statistical method called unsupervised clustering analysis to see if relatively unique groups of children could be discerned. The overall cohort successfully clustered into two groups, showing marked differences in the prevalence of respiratory infections and asthma.5 We termed the two clinical phenotypes infection and asthma prone (n = 99, 28% of the children) and non–infection and asthma prone (n = 259, 72% of the children). Infection- and asthma-prone children were significantly more likely to experience recurrent AOM, influenza, sinusitis, pneumonia, asthma, and allergic rhinitis, compared with non–infection- and asthma-prone children (Figure).
The two groups did not experience significantly different rates of eczema, food allergy, skin infections, urinary tract infections, or acute gastroenteritis, suggesting a common thread involving the respiratory tract that did not cross over to the gastrointestinal, skin, or urinary tract. We found that age at first nasopharyngeal colonization with any of the three bacterial respiratory pathogens (pneumococcus, Hflu, or Mcat) was significantly associated with the respiratory infection– and asthma-prone clinical phenotype. Specifically, respiratory infection– and asthma-prone children experienced colonization at a significantly earlier age than nonprone children did for all three bacteria. In an analysis of individual conditions, early Mcat colonization significantly associated with pneumonia, sinusitis, and asthma susceptibility; Hflu with pneumonia, sinusitis, influenza, and allergic rhinitis; and pneumococcus with sinusitis.
Since early colonization with the three bacterial respiratory pathogens was strongly associated with respiratory illnesses and asthma, nasopharyngeal microbiome analysis was performed on an available subset of samples. Bacterial diversity trended lower in infection- and asthma-prone children, consistent with dysbiosis in the respiratory infection– and asthma-prone clinical phenotype. Nine different bacteria genera were found to be differentially abundant when comparing respiratory infection– and asthma-prone and nonprone children, pointing the way to possible interventions to make the respiratory infection– and asthma-prone child nasopharyngeal microbiome more like the nonprone child.
As I have written previously in this column, recent accumulating data have shed light on the importance of the human microbiome in modulating immune homeostasis and disease susceptibility.6 My group is working toward generating new knowledge for the long-term goal of identifying new therapeutic strategies to facilitate a protective, diverse nasopharyngeal microbiome (with appropriately tuned intranasal probiotics) to prevent respiratory pathogen colonization and/or subsequent progression to respiratory infection and asthma. Also, vaccines directed against colonization-enhancing members of the microbiome may provide a means to indirectly control respiratory pathogen nasopharyngeal colonization.
Dr. Pichichero is a specialist in pediatric infectious diseases and director of the Research Institute at Rochester (N.Y.) General Hospital. He has no conflicts to declare. Contact him at [email protected]
References
1. Man WH et al. Nat Rev Microbiol. 2017;15(5):259-70.
2. Pichichero ME. J Infect. 2020;80(6):614-22.
3. Ren D et al. Clin Infect Dis. 2019;68(9):1566-74.
4. Pichichero ME et al. Pediatr Infect Dis J. 2013;32(11):1163-8.
5. Chapman T et al. PLoS One. 2020 Dec 11;15(12).
6. Blaser MJ. The microbiome revolution. J Clin Invest. 2014;124:4162-5.

