User login
Vitamin D may affect outcome in cancer patients

Cancer patients with higher levels of vitamin D at diagnosis tend to have better outcomes than patients who are vitamin D-deficient, a meta-analysis suggests.
Researchers found the association between vitamin D and prognosis was most pronounced in patients with breast cancer, colorectal cancer, and lymphomas.
The team observed an association between vitamin D and prognosis in other cancers as well, but the correlations were not significant.
Hui Wang, MD, PhD, of the Chinese Academy of Sciences in Shanghai, China, and colleagues conducted this research and reported their results in the Journal of Clinical Endocrinology & Metabolism.
The team analyzed data from 25 studies including 17,332 cancer patients. In most of the studies, patients had their vitamin D levels tested before they underwent any cancer treatment.
The analysis showed that, overall, a 10 nmol/L increase in vitamin D levels was associated with a 4% increase in survival.
The strongest links between vitamin D levels and survival were seen in breast cancer, colorectal cancer, and lymphomas. There was less evidence of a connection in patients with leukemias, lung cancer, gastric cancer, prostate cancer, melanoma, or Merkel cell carcinoma, but the available data were positive.
There were 2 studies evaluating the association between survival and vitamin D levels in leukemia patients.
Vitamin D insufficiency was associated with poor overall survival in patients with newly diagnosed chronic lymphocytic leukemia, although this was not statistically significant. The hazard ratio (HR) was 0.68 (P=0.07).
For acute myeloid leukemia, patients with vitamin D levels that were considered insufficient (20-31.9 ng/mL) or deficient (<20 ng/mL) had worse overall survival (HR=0.65) and disease-free survival (HR=0.65) than patients with normal vitamin D levels.
There were also 2 studies evaluating the association between survival and vitamin D levels in lymphoma patients. The results showed that patients with the highest vitamin D levels had better overall survival than those with the lowest vitamin D levels (HR=0.48, P<0.001).
Lymphoma patients in the highest quartile of vitamin D levels also had a lower risk of cancer-specific mortality (HR=0.50, P<0.001) and better disease-free survival (HR=0.80, P=0.04).
Similarly, higher vitamin D levels were significantly associated with reduced cancer-specific mortality for patients with colorectal cancer (P=0.005) and improved disease-free survival for patients with breast cancer (P<0.001).
For overall survival, the HR for the highest vs lowest quartile of vitamin D levels was 0.55 for colorectal cancer patients and 0.63 for breast cancer patients.
Based on these results, Dr Wang concluded, “Physicians need to pay close attention to vitamin D levels in people who have been diagnosed with cancer.” ![]()

Cancer patients with higher levels of vitamin D at diagnosis tend to have better outcomes than patients who are vitamin D-deficient, a meta-analysis suggests.
Researchers found the association between vitamin D and prognosis was most pronounced in patients with breast cancer, colorectal cancer, and lymphomas.
The team observed an association between vitamin D and prognosis in other cancers as well, but the correlations were not significant.
Hui Wang, MD, PhD, of the Chinese Academy of Sciences in Shanghai, China, and colleagues conducted this research and reported their results in the Journal of Clinical Endocrinology & Metabolism.
The team analyzed data from 25 studies including 17,332 cancer patients. In most of the studies, patients had their vitamin D levels tested before they underwent any cancer treatment.
The analysis showed that, overall, a 10 nmol/L increase in vitamin D levels was associated with a 4% increase in survival.
The strongest links between vitamin D levels and survival were seen in breast cancer, colorectal cancer, and lymphomas. There was less evidence of a connection in patients with leukemias, lung cancer, gastric cancer, prostate cancer, melanoma, or Merkel cell carcinoma, but the available data were positive.
There were 2 studies evaluating the association between survival and vitamin D levels in leukemia patients.
Vitamin D insufficiency was associated with poor overall survival in patients with newly diagnosed chronic lymphocytic leukemia, although this was not statistically significant. The hazard ratio (HR) was 0.68 (P=0.07).
For acute myeloid leukemia, patients with vitamin D levels that were considered insufficient (20-31.9 ng/mL) or deficient (<20 ng/mL) had worse overall survival (HR=0.65) and disease-free survival (HR=0.65) than patients with normal vitamin D levels.
There were also 2 studies evaluating the association between survival and vitamin D levels in lymphoma patients. The results showed that patients with the highest vitamin D levels had better overall survival than those with the lowest vitamin D levels (HR=0.48, P<0.001).
Lymphoma patients in the highest quartile of vitamin D levels also had a lower risk of cancer-specific mortality (HR=0.50, P<0.001) and better disease-free survival (HR=0.80, P=0.04).
Similarly, higher vitamin D levels were significantly associated with reduced cancer-specific mortality for patients with colorectal cancer (P=0.005) and improved disease-free survival for patients with breast cancer (P<0.001).
For overall survival, the HR for the highest vs lowest quartile of vitamin D levels was 0.55 for colorectal cancer patients and 0.63 for breast cancer patients.
Based on these results, Dr Wang concluded, “Physicians need to pay close attention to vitamin D levels in people who have been diagnosed with cancer.” ![]()

Cancer patients with higher levels of vitamin D at diagnosis tend to have better outcomes than patients who are vitamin D-deficient, a meta-analysis suggests.
Researchers found the association between vitamin D and prognosis was most pronounced in patients with breast cancer, colorectal cancer, and lymphomas.
The team observed an association between vitamin D and prognosis in other cancers as well, but the correlations were not significant.
Hui Wang, MD, PhD, of the Chinese Academy of Sciences in Shanghai, China, and colleagues conducted this research and reported their results in the Journal of Clinical Endocrinology & Metabolism.
The team analyzed data from 25 studies including 17,332 cancer patients. In most of the studies, patients had their vitamin D levels tested before they underwent any cancer treatment.
The analysis showed that, overall, a 10 nmol/L increase in vitamin D levels was associated with a 4% increase in survival.
The strongest links between vitamin D levels and survival were seen in breast cancer, colorectal cancer, and lymphomas. There was less evidence of a connection in patients with leukemias, lung cancer, gastric cancer, prostate cancer, melanoma, or Merkel cell carcinoma, but the available data were positive.
There were 2 studies evaluating the association between survival and vitamin D levels in leukemia patients.
Vitamin D insufficiency was associated with poor overall survival in patients with newly diagnosed chronic lymphocytic leukemia, although this was not statistically significant. The hazard ratio (HR) was 0.68 (P=0.07).
For acute myeloid leukemia, patients with vitamin D levels that were considered insufficient (20-31.9 ng/mL) or deficient (<20 ng/mL) had worse overall survival (HR=0.65) and disease-free survival (HR=0.65) than patients with normal vitamin D levels.
There were also 2 studies evaluating the association between survival and vitamin D levels in lymphoma patients. The results showed that patients with the highest vitamin D levels had better overall survival than those with the lowest vitamin D levels (HR=0.48, P<0.001).
Lymphoma patients in the highest quartile of vitamin D levels also had a lower risk of cancer-specific mortality (HR=0.50, P<0.001) and better disease-free survival (HR=0.80, P=0.04).
Similarly, higher vitamin D levels were significantly associated with reduced cancer-specific mortality for patients with colorectal cancer (P=0.005) and improved disease-free survival for patients with breast cancer (P<0.001).
For overall survival, the HR for the highest vs lowest quartile of vitamin D levels was 0.55 for colorectal cancer patients and 0.63 for breast cancer patients.
Based on these results, Dr Wang concluded, “Physicians need to pay close attention to vitamin D levels in people who have been diagnosed with cancer.” ![]()
AUDIO: Challenges, rewards face creators of pediatric oncofertility clinics
PHILADELPHIA – Few templates exist for creating a pediatric oncofertility clinic, particularly in the context of the nascent trend of pediatric oncologists treating adults with pediatric cancers.
Dr. Karen Burns is a pediatric oncologist at Cincinnati Children’s Hospital who was part of the team that created that institution’s own oncofertility clinic. She offered tips to others who are interested in creating a similar clinic or who would like to incorporate protocols honoring ASCO’s recent position statement that all persons with cancer deserve the right to have fertility discussed as part of their standard of care.
In this audio interview, from the annual meeting of the North American Society for Pediatric and Adolescent Gynecology, Dr. Burns addresses the challenges of treating adults with pediatric cancers, having conversations about fertility with minors, the role of advance directives in oncofertility, and what burdens oncologists with patients in these situations face and how they can be eased by working with a team.
On Twitter @whitneymcknight
PHILADELPHIA – Few templates exist for creating a pediatric oncofertility clinic, particularly in the context of the nascent trend of pediatric oncologists treating adults with pediatric cancers.
Dr. Karen Burns is a pediatric oncologist at Cincinnati Children’s Hospital who was part of the team that created that institution’s own oncofertility clinic. She offered tips to others who are interested in creating a similar clinic or who would like to incorporate protocols honoring ASCO’s recent position statement that all persons with cancer deserve the right to have fertility discussed as part of their standard of care.
In this audio interview, from the annual meeting of the North American Society for Pediatric and Adolescent Gynecology, Dr. Burns addresses the challenges of treating adults with pediatric cancers, having conversations about fertility with minors, the role of advance directives in oncofertility, and what burdens oncologists with patients in these situations face and how they can be eased by working with a team.
On Twitter @whitneymcknight
PHILADELPHIA – Few templates exist for creating a pediatric oncofertility clinic, particularly in the context of the nascent trend of pediatric oncologists treating adults with pediatric cancers.
Dr. Karen Burns is a pediatric oncologist at Cincinnati Children’s Hospital who was part of the team that created that institution’s own oncofertility clinic. She offered tips to others who are interested in creating a similar clinic or who would like to incorporate protocols honoring ASCO’s recent position statement that all persons with cancer deserve the right to have fertility discussed as part of their standard of care.
In this audio interview, from the annual meeting of the North American Society for Pediatric and Adolescent Gynecology, Dr. Burns addresses the challenges of treating adults with pediatric cancers, having conversations about fertility with minors, the role of advance directives in oncofertility, and what burdens oncologists with patients in these situations face and how they can be eased by working with a team.
On Twitter @whitneymcknight
EXPERT ANALYSIS FROM THE NASPAG ANNUAL MEETING
Team uncovers novel function of p53

Andrei Thomas Tikhonenko
Investigators have uncovered a novel role for the tumor suppressor p53, according to a paper published in Nature Cell Biology.
The research showed that loss of p53 function caused overproduction of the Aurora A kinase, an enzyme involved in cell division.
That overproduction led to mitotic spindle malformation and aberrant separation of duplicated chromosomes over daughter cells, a phenomenon that predicts tumor metastasis and poor patient outcomes.
“Attempts to identify which genetic defects drive chromosome reshuffling in human cancer led us to focus on cyclin B1 and B2, two key regulators of the stage in the cell cycle where duplicated chromosomes normally separate,” said principal investigator Jan van Deursen, PhD, of the Mayo Clinic in Rochester, Minnesota.
Dr van Deursen and his colleague, Hyun-Ja Nam, PhD, used mouse models to mimic the cyclin B1 and B2 gene defects observed in treatment-resistant human cancers. And the pair discovered that both cyclin B1 and B2 induce chromosome reshuffling and tumor formation.
Subsequent experiments investigating cyclin B2’s mechanism of action pinpointed Aurora A kinase hyperactivity as the main culprit and showed that damage or loss of p53 is a mimetic of cyclin B2 gene defects.
The investigators said the next step for this research will be testing whether anticancer drugs that inhibit Aurora A kinase can be effective in treating cancer patients whose tumors have defects in p53. ![]()

Andrei Thomas Tikhonenko
Investigators have uncovered a novel role for the tumor suppressor p53, according to a paper published in Nature Cell Biology.
The research showed that loss of p53 function caused overproduction of the Aurora A kinase, an enzyme involved in cell division.
That overproduction led to mitotic spindle malformation and aberrant separation of duplicated chromosomes over daughter cells, a phenomenon that predicts tumor metastasis and poor patient outcomes.
“Attempts to identify which genetic defects drive chromosome reshuffling in human cancer led us to focus on cyclin B1 and B2, two key regulators of the stage in the cell cycle where duplicated chromosomes normally separate,” said principal investigator Jan van Deursen, PhD, of the Mayo Clinic in Rochester, Minnesota.
Dr van Deursen and his colleague, Hyun-Ja Nam, PhD, used mouse models to mimic the cyclin B1 and B2 gene defects observed in treatment-resistant human cancers. And the pair discovered that both cyclin B1 and B2 induce chromosome reshuffling and tumor formation.
Subsequent experiments investigating cyclin B2’s mechanism of action pinpointed Aurora A kinase hyperactivity as the main culprit and showed that damage or loss of p53 is a mimetic of cyclin B2 gene defects.
The investigators said the next step for this research will be testing whether anticancer drugs that inhibit Aurora A kinase can be effective in treating cancer patients whose tumors have defects in p53. ![]()

Andrei Thomas Tikhonenko
Investigators have uncovered a novel role for the tumor suppressor p53, according to a paper published in Nature Cell Biology.
The research showed that loss of p53 function caused overproduction of the Aurora A kinase, an enzyme involved in cell division.
That overproduction led to mitotic spindle malformation and aberrant separation of duplicated chromosomes over daughter cells, a phenomenon that predicts tumor metastasis and poor patient outcomes.
“Attempts to identify which genetic defects drive chromosome reshuffling in human cancer led us to focus on cyclin B1 and B2, two key regulators of the stage in the cell cycle where duplicated chromosomes normally separate,” said principal investigator Jan van Deursen, PhD, of the Mayo Clinic in Rochester, Minnesota.
Dr van Deursen and his colleague, Hyun-Ja Nam, PhD, used mouse models to mimic the cyclin B1 and B2 gene defects observed in treatment-resistant human cancers. And the pair discovered that both cyclin B1 and B2 induce chromosome reshuffling and tumor formation.
Subsequent experiments investigating cyclin B2’s mechanism of action pinpointed Aurora A kinase hyperactivity as the main culprit and showed that damage or loss of p53 is a mimetic of cyclin B2 gene defects.
The investigators said the next step for this research will be testing whether anticancer drugs that inhibit Aurora A kinase can be effective in treating cancer patients whose tumors have defects in p53. ![]()
New insight into PTEN’s role in cancers
Researchers say they’ve uncovered new details that help explain how the PTEN gene exerts its anticancer effects and how PTEN loss or alteration can set cells on a cancerous course.
The team’s study, published in Cell, reveals that PTEN loss and PTEN mutations are not synonymous.
This discovery provides additional insight into basic tumor biology and offers a potential new direction in the pursuit of cancer therapies, according to the researchers.
“By characterizing the ways that 2 specific PTEN mutations regulate the tumor suppressor function of the normal PTEN protein, our findings suggest that different PTEN mutations contribute to tumorigenesis by regulating different aspects of PTEN biology,” said study author Pier Paolo Pandolfi, MD, PhD, of Beth Israel Deaconess Medical Center in Boston.
“It has been suggested that cancer patients harboring mutations in PTEN had poorer outcomes than cancer patients with PTEN loss. Now, using mouse modeling, we are able to demonstrate that this is indeed the case. Because PTEN mutations are extremely frequent in various types of tumors, this discovery could help pave the way for a new level of personalized cancer treatment.”
Several of the proteins that PTEN acts upon, both lipids and proteins, are known to promote cancer when bound to a phosphate. Consequently, when PTEN removes their phosphates, it is acting as a tumor suppressor to prevent cancer. When PTEN is mutated, it loses this suppressive ability, and the cancer-promoting proteins are left intact and uninhibited.
This new study showed that the PTEN mutant protein is not only functionally impaired, it also acquires the ability to affect the function of normal PTEN proteins, thereby gaining a pro-tumorigenic function. But the researchers also wanted to determine the difference between PTEN mutation and PTEN loss.
“We wanted to know, would outcomes differ in cases when PTEN was not expressed, compared with cases when PTEN was expressed but encoded a mutation within its sequence?” said study author Antonella Papa, PhD, an investigator in the Pandolfi lab.
To find out, the team created several genetically modified strains of mice to mimic the PTEN mutations found in human cancer patients.
“All mice [and humans] have 2 copies of the PTEN gene,” Dr Papa explained. “The genetically modified mice in our study had 1 copy of the PTEN gene that contained a cancer-associated mutation [either PTENC124S or PTENG129E] and 1 normal copy of PTEN. Other mice in the study had only 1 copy of the normal PTEN gene, and the second copy was removed.”
The researchers found that the mice with a single mutated copy of PTEN were more tumor-prone than the mice with a deleted copy of PTEN. They also discovered that the mutated protein that was produced by PTENC124S or PTENG129E was binding to and inhibiting the PTEN protein made from the normal copy of the PTEN gene.
“This was very surprising, as we were expecting a reduction in tumorigenesis,” Dr Papa said. “Instead, mechanistically, we found that PTEN exists as a dimer, and, in this new conformation, the mutated protein prevents the normal protein from functioning.”
At the molecular level, this generates an increased activation of a PTEN target—the protein Akt—which is what leads to the augmented tumorigenesis in the mice. Akt is part of a signaling pathway that regulates cell growth, division and metabolism.
When PTEN is prevented from inhibiting Akt, the pathway becomes overactive. As a result, targeting Akt and its pathway may be an effective treatment strategy for patients with PTEN mutations, the researchers said, adding that inhibitors to affect this pathway are currently being tested and developed.
“This defines a new working model for the function and regulation of PTEN and tells us that PTEN mutational status can be used to determine which cancer patients might benefit from earlier and more radical therapeutic interventions and, ultimately, better prognosis,” Dr Pandolfi said.
“Our findings may help to better identify and stratify patients and their response to treatment based on the different genetic alterations found in the PTEN gene. Importantly, our study shows that cancer therapy should be tailored on the basis of the very specific type of mutations that the tumor harbors.”
“This adds a new layer of complexity but also a new opportunity for precision medicine. I would say that, based on these thorough genetic analyses, this story represents the ultimate example of why personalized cancer medicine is so urgently needed.” ![]()

Researchers say they’ve uncovered new details that help explain how the PTEN gene exerts its anticancer effects and how PTEN loss or alteration can set cells on a cancerous course.
The team’s study, published in Cell, reveals that PTEN loss and PTEN mutations are not synonymous.
This discovery provides additional insight into basic tumor biology and offers a potential new direction in the pursuit of cancer therapies, according to the researchers.
“By characterizing the ways that 2 specific PTEN mutations regulate the tumor suppressor function of the normal PTEN protein, our findings suggest that different PTEN mutations contribute to tumorigenesis by regulating different aspects of PTEN biology,” said study author Pier Paolo Pandolfi, MD, PhD, of Beth Israel Deaconess Medical Center in Boston.
“It has been suggested that cancer patients harboring mutations in PTEN had poorer outcomes than cancer patients with PTEN loss. Now, using mouse modeling, we are able to demonstrate that this is indeed the case. Because PTEN mutations are extremely frequent in various types of tumors, this discovery could help pave the way for a new level of personalized cancer treatment.”
Several of the proteins that PTEN acts upon, both lipids and proteins, are known to promote cancer when bound to a phosphate. Consequently, when PTEN removes their phosphates, it is acting as a tumor suppressor to prevent cancer. When PTEN is mutated, it loses this suppressive ability, and the cancer-promoting proteins are left intact and uninhibited.
This new study showed that the PTEN mutant protein is not only functionally impaired, it also acquires the ability to affect the function of normal PTEN proteins, thereby gaining a pro-tumorigenic function. But the researchers also wanted to determine the difference between PTEN mutation and PTEN loss.
“We wanted to know, would outcomes differ in cases when PTEN was not expressed, compared with cases when PTEN was expressed but encoded a mutation within its sequence?” said study author Antonella Papa, PhD, an investigator in the Pandolfi lab.
To find out, the team created several genetically modified strains of mice to mimic the PTEN mutations found in human cancer patients.
“All mice [and humans] have 2 copies of the PTEN gene,” Dr Papa explained. “The genetically modified mice in our study had 1 copy of the PTEN gene that contained a cancer-associated mutation [either PTENC124S or PTENG129E] and 1 normal copy of PTEN. Other mice in the study had only 1 copy of the normal PTEN gene, and the second copy was removed.”
The researchers found that the mice with a single mutated copy of PTEN were more tumor-prone than the mice with a deleted copy of PTEN. They also discovered that the mutated protein that was produced by PTENC124S or PTENG129E was binding to and inhibiting the PTEN protein made from the normal copy of the PTEN gene.
“This was very surprising, as we were expecting a reduction in tumorigenesis,” Dr Papa said. “Instead, mechanistically, we found that PTEN exists as a dimer, and, in this new conformation, the mutated protein prevents the normal protein from functioning.”
At the molecular level, this generates an increased activation of a PTEN target—the protein Akt—which is what leads to the augmented tumorigenesis in the mice. Akt is part of a signaling pathway that regulates cell growth, division and metabolism.
When PTEN is prevented from inhibiting Akt, the pathway becomes overactive. As a result, targeting Akt and its pathway may be an effective treatment strategy for patients with PTEN mutations, the researchers said, adding that inhibitors to affect this pathway are currently being tested and developed.
“This defines a new working model for the function and regulation of PTEN and tells us that PTEN mutational status can be used to determine which cancer patients might benefit from earlier and more radical therapeutic interventions and, ultimately, better prognosis,” Dr Pandolfi said.
“Our findings may help to better identify and stratify patients and their response to treatment based on the different genetic alterations found in the PTEN gene. Importantly, our study shows that cancer therapy should be tailored on the basis of the very specific type of mutations that the tumor harbors.”
“This adds a new layer of complexity but also a new opportunity for precision medicine. I would say that, based on these thorough genetic analyses, this story represents the ultimate example of why personalized cancer medicine is so urgently needed.” ![]()

Researchers say they’ve uncovered new details that help explain how the PTEN gene exerts its anticancer effects and how PTEN loss or alteration can set cells on a cancerous course.
The team’s study, published in Cell, reveals that PTEN loss and PTEN mutations are not synonymous.
This discovery provides additional insight into basic tumor biology and offers a potential new direction in the pursuit of cancer therapies, according to the researchers.
“By characterizing the ways that 2 specific PTEN mutations regulate the tumor suppressor function of the normal PTEN protein, our findings suggest that different PTEN mutations contribute to tumorigenesis by regulating different aspects of PTEN biology,” said study author Pier Paolo Pandolfi, MD, PhD, of Beth Israel Deaconess Medical Center in Boston.
“It has been suggested that cancer patients harboring mutations in PTEN had poorer outcomes than cancer patients with PTEN loss. Now, using mouse modeling, we are able to demonstrate that this is indeed the case. Because PTEN mutations are extremely frequent in various types of tumors, this discovery could help pave the way for a new level of personalized cancer treatment.”
Several of the proteins that PTEN acts upon, both lipids and proteins, are known to promote cancer when bound to a phosphate. Consequently, when PTEN removes their phosphates, it is acting as a tumor suppressor to prevent cancer. When PTEN is mutated, it loses this suppressive ability, and the cancer-promoting proteins are left intact and uninhibited.
This new study showed that the PTEN mutant protein is not only functionally impaired, it also acquires the ability to affect the function of normal PTEN proteins, thereby gaining a pro-tumorigenic function. But the researchers also wanted to determine the difference between PTEN mutation and PTEN loss.
“We wanted to know, would outcomes differ in cases when PTEN was not expressed, compared with cases when PTEN was expressed but encoded a mutation within its sequence?” said study author Antonella Papa, PhD, an investigator in the Pandolfi lab.
To find out, the team created several genetically modified strains of mice to mimic the PTEN mutations found in human cancer patients.
“All mice [and humans] have 2 copies of the PTEN gene,” Dr Papa explained. “The genetically modified mice in our study had 1 copy of the PTEN gene that contained a cancer-associated mutation [either PTENC124S or PTENG129E] and 1 normal copy of PTEN. Other mice in the study had only 1 copy of the normal PTEN gene, and the second copy was removed.”
The researchers found that the mice with a single mutated copy of PTEN were more tumor-prone than the mice with a deleted copy of PTEN. They also discovered that the mutated protein that was produced by PTENC124S or PTENG129E was binding to and inhibiting the PTEN protein made from the normal copy of the PTEN gene.
“This was very surprising, as we were expecting a reduction in tumorigenesis,” Dr Papa said. “Instead, mechanistically, we found that PTEN exists as a dimer, and, in this new conformation, the mutated protein prevents the normal protein from functioning.”
At the molecular level, this generates an increased activation of a PTEN target—the protein Akt—which is what leads to the augmented tumorigenesis in the mice. Akt is part of a signaling pathway that regulates cell growth, division and metabolism.
When PTEN is prevented from inhibiting Akt, the pathway becomes overactive. As a result, targeting Akt and its pathway may be an effective treatment strategy for patients with PTEN mutations, the researchers said, adding that inhibitors to affect this pathway are currently being tested and developed.
“This defines a new working model for the function and regulation of PTEN and tells us that PTEN mutational status can be used to determine which cancer patients might benefit from earlier and more radical therapeutic interventions and, ultimately, better prognosis,” Dr Pandolfi said.
“Our findings may help to better identify and stratify patients and their response to treatment based on the different genetic alterations found in the PTEN gene. Importantly, our study shows that cancer therapy should be tailored on the basis of the very specific type of mutations that the tumor harbors.”
“This adds a new layer of complexity but also a new opportunity for precision medicine. I would say that, based on these thorough genetic analyses, this story represents the ultimate example of why personalized cancer medicine is so urgently needed.” ![]()
Group maps B-cell development
New technology has allowed scientists to create the most comprehensive map of B-cell development to date, according to a paper published in Cell.
The team combined emerging technologies for studying single cells with an advanced computational algorithm to map human B-cell development.
They believe their approach could improve researchers’ ability to investigate development in all cells and make it possible to identify rare aberrations that lead to disease.
“There are so many diseases that result from malfunctions in the molecular programs that control the development of our cell repertoire and so many rare, yet important, regulatory cell types that we have yet to discover,” said study author Dana Pe’er, PhD, of Columbia University in New York.
“We can only truly understand what goes wrong in these diseases if we have a complete map of the progression in normal development.”
Combining technologies
Dr Pe’er and her colleagues used mass cytology to observe cells in a bone marrow sample. In a single experiment, mass cytology can measure 44 molecular markers simultaneously in millions of individual cells. This provides data that can be used to compare, categorize, and order cells, as well as identify the molecular systems responsible for development.
Taking advantage of this data required the researchers to develop new mathematical and computational methods for interpreting it. Just as one can represent a physical object in 3 dimensions, the Pe’er lab’s approach involved thinking of the 44 measurements as a 44-dimensional geometric object.
So they created a new computational algorithm called Wanderlust, which uses mathematical concepts from a field called graph theory to reduce this high-dimensional data into a simple form that is easier to interpret. Wanderlust converts the developmental marker measurements in each cell into a single, 1-dimensional value that corresponds to the cell’s place within the chronology of development.
“Our body has trillions of cells of countless different types, each type bearing different molecular features and behavior,” Dr Pe’er noted. “This complexity expands from a single cell in a carefully regulated process called development.”
“This regulation creates patterns and shapes in the high-dimensional data we measure. By using Wanderlust to analyze these data, we can find the pattern and trace the trajectory that cellular development follows.”
Mapping B-cell development
To test their approach, the researchers studied development in human B cells. The team used mass cytometry to profile 44 markers in a cohort of approximately 200,000 healthy immune cells that were gathered from a single bone marrow sample.
In each cell, they measured surface markers that help identify cell type, as well as markers inside the cell that can reveal what the cell is doing, including markers for signaling, the cell cycle, apoptosis, and genome rearrangement.
Using Wanderlust to analyze the high-dimensional data provided by mass cytometry, the researchers accurately ordered the entire trajectory of 200,000 cells according to their developmental chronology. Wanderlust captured and correctly ordered all of the primary molecular landmarks known to be present in human B-cell development.
The algorithm also pinpointed a number of previously unknown regulatory signaling checkpoints that are required for human B-cell development, as well as uncharacterized subtypes of B-cell progenitors that correspond to developmental stages.
The researchers identified rare, previously unknown signaling events involving STAT5 that occurred in just 7 out of 10,000 cells. The team found that disrupting these signaling events using kinase inhibitors fully stalled the development of B cells.
Identifying and characterizing the regulatory checkpoints that control and monitor cell fate can have many practical applications, the researchers said, including the development of new diagnostics and therapeutics.
Furthermore, the team’s mapping process can be applied to any type of cell. They believe their method offers the possibility of studying normal development as well as the processes responsible for any kind of developmental disease.
“This current project is a landmark, both in the study of development and in single-cell research, and has completely changed the way I think about science,” Dr Pe’er said. “A fire has been lit, and these findings are just the tip of the iceberg of what is now possible.” ![]()

New technology has allowed scientists to create the most comprehensive map of B-cell development to date, according to a paper published in Cell.
The team combined emerging technologies for studying single cells with an advanced computational algorithm to map human B-cell development.
They believe their approach could improve researchers’ ability to investigate development in all cells and make it possible to identify rare aberrations that lead to disease.
“There are so many diseases that result from malfunctions in the molecular programs that control the development of our cell repertoire and so many rare, yet important, regulatory cell types that we have yet to discover,” said study author Dana Pe’er, PhD, of Columbia University in New York.
“We can only truly understand what goes wrong in these diseases if we have a complete map of the progression in normal development.”
Combining technologies
Dr Pe’er and her colleagues used mass cytology to observe cells in a bone marrow sample. In a single experiment, mass cytology can measure 44 molecular markers simultaneously in millions of individual cells. This provides data that can be used to compare, categorize, and order cells, as well as identify the molecular systems responsible for development.
Taking advantage of this data required the researchers to develop new mathematical and computational methods for interpreting it. Just as one can represent a physical object in 3 dimensions, the Pe’er lab’s approach involved thinking of the 44 measurements as a 44-dimensional geometric object.
So they created a new computational algorithm called Wanderlust, which uses mathematical concepts from a field called graph theory to reduce this high-dimensional data into a simple form that is easier to interpret. Wanderlust converts the developmental marker measurements in each cell into a single, 1-dimensional value that corresponds to the cell’s place within the chronology of development.
“Our body has trillions of cells of countless different types, each type bearing different molecular features and behavior,” Dr Pe’er noted. “This complexity expands from a single cell in a carefully regulated process called development.”
“This regulation creates patterns and shapes in the high-dimensional data we measure. By using Wanderlust to analyze these data, we can find the pattern and trace the trajectory that cellular development follows.”
Mapping B-cell development
To test their approach, the researchers studied development in human B cells. The team used mass cytometry to profile 44 markers in a cohort of approximately 200,000 healthy immune cells that were gathered from a single bone marrow sample.
In each cell, they measured surface markers that help identify cell type, as well as markers inside the cell that can reveal what the cell is doing, including markers for signaling, the cell cycle, apoptosis, and genome rearrangement.
Using Wanderlust to analyze the high-dimensional data provided by mass cytometry, the researchers accurately ordered the entire trajectory of 200,000 cells according to their developmental chronology. Wanderlust captured and correctly ordered all of the primary molecular landmarks known to be present in human B-cell development.
The algorithm also pinpointed a number of previously unknown regulatory signaling checkpoints that are required for human B-cell development, as well as uncharacterized subtypes of B-cell progenitors that correspond to developmental stages.
The researchers identified rare, previously unknown signaling events involving STAT5 that occurred in just 7 out of 10,000 cells. The team found that disrupting these signaling events using kinase inhibitors fully stalled the development of B cells.
Identifying and characterizing the regulatory checkpoints that control and monitor cell fate can have many practical applications, the researchers said, including the development of new diagnostics and therapeutics.
Furthermore, the team’s mapping process can be applied to any type of cell. They believe their method offers the possibility of studying normal development as well as the processes responsible for any kind of developmental disease.
“This current project is a landmark, both in the study of development and in single-cell research, and has completely changed the way I think about science,” Dr Pe’er said. “A fire has been lit, and these findings are just the tip of the iceberg of what is now possible.” ![]()

New technology has allowed scientists to create the most comprehensive map of B-cell development to date, according to a paper published in Cell.
The team combined emerging technologies for studying single cells with an advanced computational algorithm to map human B-cell development.
They believe their approach could improve researchers’ ability to investigate development in all cells and make it possible to identify rare aberrations that lead to disease.
“There are so many diseases that result from malfunctions in the molecular programs that control the development of our cell repertoire and so many rare, yet important, regulatory cell types that we have yet to discover,” said study author Dana Pe’er, PhD, of Columbia University in New York.
“We can only truly understand what goes wrong in these diseases if we have a complete map of the progression in normal development.”
Combining technologies
Dr Pe’er and her colleagues used mass cytology to observe cells in a bone marrow sample. In a single experiment, mass cytology can measure 44 molecular markers simultaneously in millions of individual cells. This provides data that can be used to compare, categorize, and order cells, as well as identify the molecular systems responsible for development.
Taking advantage of this data required the researchers to develop new mathematical and computational methods for interpreting it. Just as one can represent a physical object in 3 dimensions, the Pe’er lab’s approach involved thinking of the 44 measurements as a 44-dimensional geometric object.
So they created a new computational algorithm called Wanderlust, which uses mathematical concepts from a field called graph theory to reduce this high-dimensional data into a simple form that is easier to interpret. Wanderlust converts the developmental marker measurements in each cell into a single, 1-dimensional value that corresponds to the cell’s place within the chronology of development.
“Our body has trillions of cells of countless different types, each type bearing different molecular features and behavior,” Dr Pe’er noted. “This complexity expands from a single cell in a carefully regulated process called development.”
“This regulation creates patterns and shapes in the high-dimensional data we measure. By using Wanderlust to analyze these data, we can find the pattern and trace the trajectory that cellular development follows.”
Mapping B-cell development
To test their approach, the researchers studied development in human B cells. The team used mass cytometry to profile 44 markers in a cohort of approximately 200,000 healthy immune cells that were gathered from a single bone marrow sample.
In each cell, they measured surface markers that help identify cell type, as well as markers inside the cell that can reveal what the cell is doing, including markers for signaling, the cell cycle, apoptosis, and genome rearrangement.
Using Wanderlust to analyze the high-dimensional data provided by mass cytometry, the researchers accurately ordered the entire trajectory of 200,000 cells according to their developmental chronology. Wanderlust captured and correctly ordered all of the primary molecular landmarks known to be present in human B-cell development.
The algorithm also pinpointed a number of previously unknown regulatory signaling checkpoints that are required for human B-cell development, as well as uncharacterized subtypes of B-cell progenitors that correspond to developmental stages.
The researchers identified rare, previously unknown signaling events involving STAT5 that occurred in just 7 out of 10,000 cells. The team found that disrupting these signaling events using kinase inhibitors fully stalled the development of B cells.
Identifying and characterizing the regulatory checkpoints that control and monitor cell fate can have many practical applications, the researchers said, including the development of new diagnostics and therapeutics.
Furthermore, the team’s mapping process can be applied to any type of cell. They believe their method offers the possibility of studying normal development as well as the processes responsible for any kind of developmental disease.
“This current project is a landmark, both in the study of development and in single-cell research, and has completely changed the way I think about science,” Dr Pe’er said. “A fire has been lit, and these findings are just the tip of the iceberg of what is now possible.” ![]()
FDA approves first drug for multicentric Castleman’s disease
Credit: Janssen Biotech, Inc.
The US Food and Drug Administration (FDA) has authorized marketing of siltuximab (Sylvant), the first drug approved to treat patients with multicentric Castleman’s disease (MCD) who are negative for human immunodeficiency virus (HIV) and human herpes virus 8 (HHV-8).
Siltuximab is a chimeric monoclonal antibody that binds to interleukin-6 (IL-6). Dysregulated overproduction of IL-6 has been implicated in the pathogenesis of MCD.
Siltuximab has not been studied in MCD patients who are HIV- or HHV-8 positive because the drug did not bind to virally produced IL-6 in a nonclinical study.
MCD is a rare blood disorder in which lymphocytes are overproduced, leading to enlarged lymph nodes. MCD can also affect lymphoid tissue of internal organs, causing enlargement of the liver, spleen, or other organs.
Patients with MCD have a high risk of death. Infections, multisystem organ failure, and malignancies such as lymphoma are common causes of death in patients with MCD.
“There has been a serious need for treatment options for patients with MCD,” said Frits van Rhee, MD, PhD, a professor at the University of Arkansas for Medical Sciences and lead investigator of the MCD2001 study.
“MCD is a complex disease, and, up until this point, physicians have tried to reduce lymph node masses and put the disease in remission through a combination of treatments, but MCD often returns. [The approval of siltuximab] gives physicians a long-awaited treatment option for [patients] suffering with this chronic, serious, and debilitating disease.”
The FDA reviewed siltuximab under its priority review program, which provides an expedited review for drugs that demonstrate the potential to be a significant improvement in safety or effectiveness in the treatment of a serious condition. The drug was also granted orphan designation, as it is intended to treat a rare disease.
MCD2001: A phase 2 study of siltuximab
The FDA approval of siltuximab is based on results of the phase 2 MCD2001 trial. This randomized, double-blind, placebo-controlled study enrolled 79 patients with symptomatic MCD that was HIV- and HHV-8 negative.
Fifty-three patients were randomized to receive siltuximab at a dose of 11 mg/kg, plus best supportive care. The remaining 26 patients were randomized to receive placebo plus best supportive care.
The researchers defined a durable response as a tumor and symptomatic response that persisted for a minimum of 18 weeks without treatment failure. Thirty-four percent of patients in the siltuximab arm achieved this endpoint, but none of the patients in the placebo arm did (P=0.0012).
On the other hand, 4% of patients in the placebo arm experienced a tumor response, as did 38% of patients in the siltuximab arm (P<0.05).
The median time to treatment failure was not reached for patients in the siltuximab arm. But patients in the placebo arm experienced treatment failure at a median of 134 days (P<0.05).
The most frequent adverse events in siltuximab-treated patients (greater than 10% compared to placebo) were rash (28%), pruritus (28%), upper respiratory tract infection (26%), weight gain (19%), and hyperuricemia (11%).
Results of this study were presented at the 2013 ASH Annual Meeting and published in Blood. The study was sponsored by Janssen Research & Development, the company developing siltuximab.
Access to siltuximab
Siltuximab is marketed as Sylvant by Janssen Biotech Inc., which is based in Horsham, Pennsylvania. To promote access to the drug, Janssen has created the SylvantOne™ Support program.
The program offers services for providers and patients that can help assess insurance coverage and identify cost support options, such as the SylvantOne™ Patient Rebate Program for eligible commercial patients, as well as a potential option for those who are uninsured.
Patients and providers can contact SylvantOne™ Support by calling 1-855-299-8844.
Siltuximab is available in a 100-mg, single-use vial of lyophilized powder and a 400-mg, single-use vial of lyophilized powder. The recommended dose of siltuximab is 11 mg/kg given over 1 hour, via intravenous infusion, every 3 weeks.
For more information on siltuximab, see the full prescribing information. ![]()

Credit: Janssen Biotech, Inc.
The US Food and Drug Administration (FDA) has authorized marketing of siltuximab (Sylvant), the first drug approved to treat patients with multicentric Castleman’s disease (MCD) who are negative for human immunodeficiency virus (HIV) and human herpes virus 8 (HHV-8).
Siltuximab is a chimeric monoclonal antibody that binds to interleukin-6 (IL-6). Dysregulated overproduction of IL-6 has been implicated in the pathogenesis of MCD.
Siltuximab has not been studied in MCD patients who are HIV- or HHV-8 positive because the drug did not bind to virally produced IL-6 in a nonclinical study.
MCD is a rare blood disorder in which lymphocytes are overproduced, leading to enlarged lymph nodes. MCD can also affect lymphoid tissue of internal organs, causing enlargement of the liver, spleen, or other organs.
Patients with MCD have a high risk of death. Infections, multisystem organ failure, and malignancies such as lymphoma are common causes of death in patients with MCD.
“There has been a serious need for treatment options for patients with MCD,” said Frits van Rhee, MD, PhD, a professor at the University of Arkansas for Medical Sciences and lead investigator of the MCD2001 study.
“MCD is a complex disease, and, up until this point, physicians have tried to reduce lymph node masses and put the disease in remission through a combination of treatments, but MCD often returns. [The approval of siltuximab] gives physicians a long-awaited treatment option for [patients] suffering with this chronic, serious, and debilitating disease.”
The FDA reviewed siltuximab under its priority review program, which provides an expedited review for drugs that demonstrate the potential to be a significant improvement in safety or effectiveness in the treatment of a serious condition. The drug was also granted orphan designation, as it is intended to treat a rare disease.
MCD2001: A phase 2 study of siltuximab
The FDA approval of siltuximab is based on results of the phase 2 MCD2001 trial. This randomized, double-blind, placebo-controlled study enrolled 79 patients with symptomatic MCD that was HIV- and HHV-8 negative.
Fifty-three patients were randomized to receive siltuximab at a dose of 11 mg/kg, plus best supportive care. The remaining 26 patients were randomized to receive placebo plus best supportive care.
The researchers defined a durable response as a tumor and symptomatic response that persisted for a minimum of 18 weeks without treatment failure. Thirty-four percent of patients in the siltuximab arm achieved this endpoint, but none of the patients in the placebo arm did (P=0.0012).
On the other hand, 4% of patients in the placebo arm experienced a tumor response, as did 38% of patients in the siltuximab arm (P<0.05).
The median time to treatment failure was not reached for patients in the siltuximab arm. But patients in the placebo arm experienced treatment failure at a median of 134 days (P<0.05).
The most frequent adverse events in siltuximab-treated patients (greater than 10% compared to placebo) were rash (28%), pruritus (28%), upper respiratory tract infection (26%), weight gain (19%), and hyperuricemia (11%).
Results of this study were presented at the 2013 ASH Annual Meeting and published in Blood. The study was sponsored by Janssen Research & Development, the company developing siltuximab.
Access to siltuximab
Siltuximab is marketed as Sylvant by Janssen Biotech Inc., which is based in Horsham, Pennsylvania. To promote access to the drug, Janssen has created the SylvantOne™ Support program.
The program offers services for providers and patients that can help assess insurance coverage and identify cost support options, such as the SylvantOne™ Patient Rebate Program for eligible commercial patients, as well as a potential option for those who are uninsured.
Patients and providers can contact SylvantOne™ Support by calling 1-855-299-8844.
Siltuximab is available in a 100-mg, single-use vial of lyophilized powder and a 400-mg, single-use vial of lyophilized powder. The recommended dose of siltuximab is 11 mg/kg given over 1 hour, via intravenous infusion, every 3 weeks.
For more information on siltuximab, see the full prescribing information. ![]()

Credit: Janssen Biotech, Inc.
The US Food and Drug Administration (FDA) has authorized marketing of siltuximab (Sylvant), the first drug approved to treat patients with multicentric Castleman’s disease (MCD) who are negative for human immunodeficiency virus (HIV) and human herpes virus 8 (HHV-8).
Siltuximab is a chimeric monoclonal antibody that binds to interleukin-6 (IL-6). Dysregulated overproduction of IL-6 has been implicated in the pathogenesis of MCD.
Siltuximab has not been studied in MCD patients who are HIV- or HHV-8 positive because the drug did not bind to virally produced IL-6 in a nonclinical study.
MCD is a rare blood disorder in which lymphocytes are overproduced, leading to enlarged lymph nodes. MCD can also affect lymphoid tissue of internal organs, causing enlargement of the liver, spleen, or other organs.
Patients with MCD have a high risk of death. Infections, multisystem organ failure, and malignancies such as lymphoma are common causes of death in patients with MCD.
“There has been a serious need for treatment options for patients with MCD,” said Frits van Rhee, MD, PhD, a professor at the University of Arkansas for Medical Sciences and lead investigator of the MCD2001 study.
“MCD is a complex disease, and, up until this point, physicians have tried to reduce lymph node masses and put the disease in remission through a combination of treatments, but MCD often returns. [The approval of siltuximab] gives physicians a long-awaited treatment option for [patients] suffering with this chronic, serious, and debilitating disease.”
The FDA reviewed siltuximab under its priority review program, which provides an expedited review for drugs that demonstrate the potential to be a significant improvement in safety or effectiveness in the treatment of a serious condition. The drug was also granted orphan designation, as it is intended to treat a rare disease.
MCD2001: A phase 2 study of siltuximab
The FDA approval of siltuximab is based on results of the phase 2 MCD2001 trial. This randomized, double-blind, placebo-controlled study enrolled 79 patients with symptomatic MCD that was HIV- and HHV-8 negative.
Fifty-three patients were randomized to receive siltuximab at a dose of 11 mg/kg, plus best supportive care. The remaining 26 patients were randomized to receive placebo plus best supportive care.
The researchers defined a durable response as a tumor and symptomatic response that persisted for a minimum of 18 weeks without treatment failure. Thirty-four percent of patients in the siltuximab arm achieved this endpoint, but none of the patients in the placebo arm did (P=0.0012).
On the other hand, 4% of patients in the placebo arm experienced a tumor response, as did 38% of patients in the siltuximab arm (P<0.05).
The median time to treatment failure was not reached for patients in the siltuximab arm. But patients in the placebo arm experienced treatment failure at a median of 134 days (P<0.05).
The most frequent adverse events in siltuximab-treated patients (greater than 10% compared to placebo) were rash (28%), pruritus (28%), upper respiratory tract infection (26%), weight gain (19%), and hyperuricemia (11%).
Results of this study were presented at the 2013 ASH Annual Meeting and published in Blood. The study was sponsored by Janssen Research & Development, the company developing siltuximab.
Access to siltuximab
Siltuximab is marketed as Sylvant by Janssen Biotech Inc., which is based in Horsham, Pennsylvania. To promote access to the drug, Janssen has created the SylvantOne™ Support program.
The program offers services for providers and patients that can help assess insurance coverage and identify cost support options, such as the SylvantOne™ Patient Rebate Program for eligible commercial patients, as well as a potential option for those who are uninsured.
Patients and providers can contact SylvantOne™ Support by calling 1-855-299-8844.
Siltuximab is available in a 100-mg, single-use vial of lyophilized powder and a 400-mg, single-use vial of lyophilized powder. The recommended dose of siltuximab is 11 mg/kg given over 1 hour, via intravenous infusion, every 3 weeks.
For more information on siltuximab, see the full prescribing information. ![]()
Leukemic breast tumors may cause resistance in AML, ALL
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
Protein helps HSP90 inhibitors fight cancers
Credit: PNAS
Researchers have discovered how a molecule called CUL5 helps HSP90 inhibitors kill cancer cells, according to a study published in Proceedings of the National Academy of Sciences.
The team found that CUL5 is required for the degradation of proteins that promote cancer cell proliferation, and CUL5 works in opposition to HSP90.
When cancer cells are treated with HSP90 inhibitors, CUL5 immediately steps in to help dispose of the proliferation-promoting proteins.
Based on these findings, the researchers speculate that some patients may be resistant to HSP90 inhibitors if their cancer cells have lower amounts of CUL5. And conversely, the drugs may work better in patients with higher CUL5 levels.
Paul Workman, PhD, of The Institute of Cancer Research in London, UK, and his colleagues conducted this research in cell lines of melanoma, as well as colon, breast, and lung cancers.
They first tested the HSP90 inhibitor 17-AAG in HT29 cells and found that CUL5 is involved in the drug-induced degradation of several protein kinase clients of HSP90.
Then, the researchers assessed the effects of silencing CUL5 and discovered that it delays the abrogation of protein signaling caused by an HSP90 inhibitor.
Furthermore, silencing CUL5 reduced cellular sensitivity to 3 different HSP90 inhibitors across the 4 different cancer types studied, which, as the researchers pointed out, are driven by different protein kinases.
So the team believes this research could apply to a number of different cancers. HSP90 inhibitors have proven effective against a range of malignancies, including leukemias, lymphomas, and multiple myeloma.
“We’ve known for some time that drugs that block HSP90 have great potential as treatments for cancers . . . , and we had an initial clue that the protein CUL5 may be involved in some way in how these drugs work,” Dr Workman said.
“Our new research shows that CUL5 is not only vital in the response of cancer cells to HSP90 inhibitors but also reveals surprising insights into precisely how it works by acting at several different levels. What also surprised us was that CUL5 gets rid of many more of the cancer-causing proteins than we’d previously imagined and that it’s effective across several types of tumor.”
“This suggests that a test for CUL5 in patients could help us tell whether they might respond to HSP90-blocking drugs, as well as pointing to new targets to develop more effective drugs.” ![]()

Credit: PNAS
Researchers have discovered how a molecule called CUL5 helps HSP90 inhibitors kill cancer cells, according to a study published in Proceedings of the National Academy of Sciences.
The team found that CUL5 is required for the degradation of proteins that promote cancer cell proliferation, and CUL5 works in opposition to HSP90.
When cancer cells are treated with HSP90 inhibitors, CUL5 immediately steps in to help dispose of the proliferation-promoting proteins.
Based on these findings, the researchers speculate that some patients may be resistant to HSP90 inhibitors if their cancer cells have lower amounts of CUL5. And conversely, the drugs may work better in patients with higher CUL5 levels.
Paul Workman, PhD, of The Institute of Cancer Research in London, UK, and his colleagues conducted this research in cell lines of melanoma, as well as colon, breast, and lung cancers.
They first tested the HSP90 inhibitor 17-AAG in HT29 cells and found that CUL5 is involved in the drug-induced degradation of several protein kinase clients of HSP90.
Then, the researchers assessed the effects of silencing CUL5 and discovered that it delays the abrogation of protein signaling caused by an HSP90 inhibitor.
Furthermore, silencing CUL5 reduced cellular sensitivity to 3 different HSP90 inhibitors across the 4 different cancer types studied, which, as the researchers pointed out, are driven by different protein kinases.
So the team believes this research could apply to a number of different cancers. HSP90 inhibitors have proven effective against a range of malignancies, including leukemias, lymphomas, and multiple myeloma.
“We’ve known for some time that drugs that block HSP90 have great potential as treatments for cancers . . . , and we had an initial clue that the protein CUL5 may be involved in some way in how these drugs work,” Dr Workman said.
“Our new research shows that CUL5 is not only vital in the response of cancer cells to HSP90 inhibitors but also reveals surprising insights into precisely how it works by acting at several different levels. What also surprised us was that CUL5 gets rid of many more of the cancer-causing proteins than we’d previously imagined and that it’s effective across several types of tumor.”
“This suggests that a test for CUL5 in patients could help us tell whether they might respond to HSP90-blocking drugs, as well as pointing to new targets to develop more effective drugs.” ![]()

Credit: PNAS
Researchers have discovered how a molecule called CUL5 helps HSP90 inhibitors kill cancer cells, according to a study published in Proceedings of the National Academy of Sciences.
The team found that CUL5 is required for the degradation of proteins that promote cancer cell proliferation, and CUL5 works in opposition to HSP90.
When cancer cells are treated with HSP90 inhibitors, CUL5 immediately steps in to help dispose of the proliferation-promoting proteins.
Based on these findings, the researchers speculate that some patients may be resistant to HSP90 inhibitors if their cancer cells have lower amounts of CUL5. And conversely, the drugs may work better in patients with higher CUL5 levels.
Paul Workman, PhD, of The Institute of Cancer Research in London, UK, and his colleagues conducted this research in cell lines of melanoma, as well as colon, breast, and lung cancers.
They first tested the HSP90 inhibitor 17-AAG in HT29 cells and found that CUL5 is involved in the drug-induced degradation of several protein kinase clients of HSP90.
Then, the researchers assessed the effects of silencing CUL5 and discovered that it delays the abrogation of protein signaling caused by an HSP90 inhibitor.
Furthermore, silencing CUL5 reduced cellular sensitivity to 3 different HSP90 inhibitors across the 4 different cancer types studied, which, as the researchers pointed out, are driven by different protein kinases.
So the team believes this research could apply to a number of different cancers. HSP90 inhibitors have proven effective against a range of malignancies, including leukemias, lymphomas, and multiple myeloma.
“We’ve known for some time that drugs that block HSP90 have great potential as treatments for cancers . . . , and we had an initial clue that the protein CUL5 may be involved in some way in how these drugs work,” Dr Workman said.
“Our new research shows that CUL5 is not only vital in the response of cancer cells to HSP90 inhibitors but also reveals surprising insights into precisely how it works by acting at several different levels. What also surprised us was that CUL5 gets rid of many more of the cancer-causing proteins than we’d previously imagined and that it’s effective across several types of tumor.”
“This suggests that a test for CUL5 in patients could help us tell whether they might respond to HSP90-blocking drugs, as well as pointing to new targets to develop more effective drugs.”
Molecule shows preclinical activity in leukemias, lymphomas
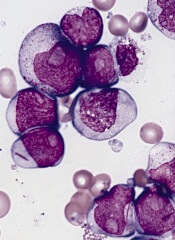
SAN DIEGO—A small molecule that has previously proven effective against solid tumors exhibits activity against leukemias and lymphomas, preclinical research suggests.
The molecule, LOR-253, showed antiproliferative activity in a range of leukemia and lymphoma cell lines, induced apoptosis in acute myeloid leukemia (AML) in vitro, and demonstrated synergy with chemotherapeutic agents.
Ronnie Lum, PhD, and colleagues at Lorus Therapeutics, Inc., the Toronto, Canada-based company developing LOR-253, presented these results at the AACR Annual Meeting 2014 (abstract 4544).
LOR-253 acts through induction of the innate tumor suppressor KLF4. Recent research has suggested that upregulation of the transcription factor CDX2 drives the development or progression of leukemic disease. And CDX2 has been shown to silence KLF4, which is reported to be a critical oncogenic event in AML.
Wih this in mind, the researchers decided to test LOR-253’s activity against AML and other hematologic malignancies in vitro.
Experiments revealed that LOR-253 exerts antiproliferative activity against a range of leukemia and lymphoma cell lines, including Ramos, Raji, K-562, Jurkat, MOLT-4, CCRF-CEM, HEL92.1.7, MOLM-13, THP-1, MV411, NB4, HL-60, KG-1, NOMO-1, SKM-1, OCI-AML-2, EOL-1, and Kasumi-1.
IC50 values were substantially lower in these cell lines than in melanoma cell lines, as well as lines of lung, bladder, colon, prostate, and breast cancers.
The researchers also found that LOR-253 induces KLF4 mRNA expression in the AML cell lines HL60 and THP1. This prompts increased expression of p21, a cyclin-dependent kinase inhibitor that is transcriptionally regulated by KLF4.
Consistent with these results, LOR-253 induced cell-cycle arrest and apoptosis in the AML cell lines, which suggests the molecule acted through its intended mechanism of action.
LOR-253 also showed “strong anticancer synergy” in HL60 cells when delivered in combination with daunorubicin, azacitidine, decitabine, or cytarabine.
When LOR-253 was delivered concurrently with chemotherapy, cell viability decreased the most with cytarabine, followed by decitabine, azacitidine, and daunorubicin. With sequential treatment, cell viability decreased the most when LOR-253 was delivered with decitabine, followed by azacitidine, cytarabine, and daunorubicin.
The researchers said these results suggest LOR-253 could provide a new approach to treat AML and, possibly, other hematologic malignancies.
They are now conducting studies to further characterize the pathway that mediates KLF4 induction by LOR-253, to characterize the effects of LOR-253 in combination with approved chemotherapies for AML, and to assess the efficacy of LOR-253 in animal models of AML.
Lorus Therapeutics is also planning a dose-escalating, phase 1b trial of LOR-253 as monotherapy in AML, myelodysplastic syndromes, and other hematologic malignancies. The company expects to begin the trial this summer.

SAN DIEGO—A small molecule that has previously proven effective against solid tumors exhibits activity against leukemias and lymphomas, preclinical research suggests.
The molecule, LOR-253, showed antiproliferative activity in a range of leukemia and lymphoma cell lines, induced apoptosis in acute myeloid leukemia (AML) in vitro, and demonstrated synergy with chemotherapeutic agents.
Ronnie Lum, PhD, and colleagues at Lorus Therapeutics, Inc., the Toronto, Canada-based company developing LOR-253, presented these results at the AACR Annual Meeting 2014 (abstract 4544).
LOR-253 acts through induction of the innate tumor suppressor KLF4. Recent research has suggested that upregulation of the transcription factor CDX2 drives the development or progression of leukemic disease. And CDX2 has been shown to silence KLF4, which is reported to be a critical oncogenic event in AML.
Wih this in mind, the researchers decided to test LOR-253’s activity against AML and other hematologic malignancies in vitro.
Experiments revealed that LOR-253 exerts antiproliferative activity against a range of leukemia and lymphoma cell lines, including Ramos, Raji, K-562, Jurkat, MOLT-4, CCRF-CEM, HEL92.1.7, MOLM-13, THP-1, MV411, NB4, HL-60, KG-1, NOMO-1, SKM-1, OCI-AML-2, EOL-1, and Kasumi-1.
IC50 values were substantially lower in these cell lines than in melanoma cell lines, as well as lines of lung, bladder, colon, prostate, and breast cancers.
The researchers also found that LOR-253 induces KLF4 mRNA expression in the AML cell lines HL60 and THP1. This prompts increased expression of p21, a cyclin-dependent kinase inhibitor that is transcriptionally regulated by KLF4.
Consistent with these results, LOR-253 induced cell-cycle arrest and apoptosis in the AML cell lines, which suggests the molecule acted through its intended mechanism of action.
LOR-253 also showed “strong anticancer synergy” in HL60 cells when delivered in combination with daunorubicin, azacitidine, decitabine, or cytarabine.
When LOR-253 was delivered concurrently with chemotherapy, cell viability decreased the most with cytarabine, followed by decitabine, azacitidine, and daunorubicin. With sequential treatment, cell viability decreased the most when LOR-253 was delivered with decitabine, followed by azacitidine, cytarabine, and daunorubicin.
The researchers said these results suggest LOR-253 could provide a new approach to treat AML and, possibly, other hematologic malignancies.
They are now conducting studies to further characterize the pathway that mediates KLF4 induction by LOR-253, to characterize the effects of LOR-253 in combination with approved chemotherapies for AML, and to assess the efficacy of LOR-253 in animal models of AML.
Lorus Therapeutics is also planning a dose-escalating, phase 1b trial of LOR-253 as monotherapy in AML, myelodysplastic syndromes, and other hematologic malignancies. The company expects to begin the trial this summer.

SAN DIEGO—A small molecule that has previously proven effective against solid tumors exhibits activity against leukemias and lymphomas, preclinical research suggests.
The molecule, LOR-253, showed antiproliferative activity in a range of leukemia and lymphoma cell lines, induced apoptosis in acute myeloid leukemia (AML) in vitro, and demonstrated synergy with chemotherapeutic agents.
Ronnie Lum, PhD, and colleagues at Lorus Therapeutics, Inc., the Toronto, Canada-based company developing LOR-253, presented these results at the AACR Annual Meeting 2014 (abstract 4544).
LOR-253 acts through induction of the innate tumor suppressor KLF4. Recent research has suggested that upregulation of the transcription factor CDX2 drives the development or progression of leukemic disease. And CDX2 has been shown to silence KLF4, which is reported to be a critical oncogenic event in AML.
Wih this in mind, the researchers decided to test LOR-253’s activity against AML and other hematologic malignancies in vitro.
Experiments revealed that LOR-253 exerts antiproliferative activity against a range of leukemia and lymphoma cell lines, including Ramos, Raji, K-562, Jurkat, MOLT-4, CCRF-CEM, HEL92.1.7, MOLM-13, THP-1, MV411, NB4, HL-60, KG-1, NOMO-1, SKM-1, OCI-AML-2, EOL-1, and Kasumi-1.
IC50 values were substantially lower in these cell lines than in melanoma cell lines, as well as lines of lung, bladder, colon, prostate, and breast cancers.
The researchers also found that LOR-253 induces KLF4 mRNA expression in the AML cell lines HL60 and THP1. This prompts increased expression of p21, a cyclin-dependent kinase inhibitor that is transcriptionally regulated by KLF4.
Consistent with these results, LOR-253 induced cell-cycle arrest and apoptosis in the AML cell lines, which suggests the molecule acted through its intended mechanism of action.
LOR-253 also showed “strong anticancer synergy” in HL60 cells when delivered in combination with daunorubicin, azacitidine, decitabine, or cytarabine.
When LOR-253 was delivered concurrently with chemotherapy, cell viability decreased the most with cytarabine, followed by decitabine, azacitidine, and daunorubicin. With sequential treatment, cell viability decreased the most when LOR-253 was delivered with decitabine, followed by azacitidine, cytarabine, and daunorubicin.
The researchers said these results suggest LOR-253 could provide a new approach to treat AML and, possibly, other hematologic malignancies.
They are now conducting studies to further characterize the pathway that mediates KLF4 induction by LOR-253, to characterize the effects of LOR-253 in combination with approved chemotherapies for AML, and to assess the efficacy of LOR-253 in animal models of AML.
Lorus Therapeutics is also planning a dose-escalating, phase 1b trial of LOR-253 as monotherapy in AML, myelodysplastic syndromes, and other hematologic malignancies. The company expects to begin the trial this summer.
Compound targets mutated DLBCL, WM cells
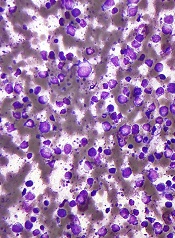
SAN DIEGO—A Toll-like receptor (TLR) antagonist can target B-cell lymphoma cells harboring the MYD88 L265P mutation, preclinical research suggests.
The compound, IMO-8400, decreased the viability of mutated diffuse large B-cell lymphoma (DLBCL) cells and Waldenström’s macroglobulinemia (WM) cells in vitro.
IMO-8400 also decreased tumor growth and prolonged survival in mice with MYD88 L265P-positive DLBCL.
Lakshmi Bhagat, PhD, and colleagues from the Cambridge, Massachusetts-based Idera Pharmaceuticals, Inc.—the company developing IMO-8400—presented these results at the AACR Annual Meeting 2014 (abstract 2570).
The researchers said their data provide additional evidence that the MYD88 L265P mutation results in over-activation of TLR7- and TLR9-mediated signaling, and blocking these TLRs leads to tumor cell death. IMO-8400 is an oligonucleotide-based antagonist of TLRs 7, 8, and 9.
In experiments with OCI‐Ly10 cells (DLBCL cells harboring the MYD88 L265P mutation), IMO-8400 prompted cell death and decreased proliferative cell signaling. But the compound did not produce these effects in SU-DHL-6 cells (DLBCL cells without the MYD88 L265P mutation).
In OCI‐Ly10 cells, IMO-8400 inhibited the IRAK-1, IRAK-4, BTK, STAT-3, Ik-Ba, and NF-κB pathways. The compound did not affect signaling pathways in SU-DHL-6 cells.
IMO-8400 also inhibited tumor growth in a mouse model of MYD88 L265P-positive, activated B-cell-like DLBCL. This inhibition was linked to the suppression of tumor-associated cytokines, including human IL-10, IL-2R, IP-10, and MIG.
Treated mice had significantly longer survival than controls, and the effect was dose-dependent. When IMO-8400 was given at 12.5 mg/kg, the P value was 0.0002. At 25 mg/kg, the P value was less than 0.0002. And at 50 mg/kg, the P value was less than 0.0001.
The researchers also found that IMO‐8400 inhibited cell viability, cytokine production, and signaling pathways in MYD88 L265P-positive WM cells. They observed these effects in the MWCL‐1 cell line and in cells from WM patients.
The team said these results provide a “strong foundation” for accelerating the clinical development of IMO-8400 in patients with B-cell lymphomas harboring the MYD88 L265P mutation.
To that end, Idera has opened enrollment in a phase 1/2 trial of IMO-8400 in WM patients who are refractory to prior therapies. The company has also submitted a protocol to the US Food and Drug Administration to conduct a phase 1/2 trial in patients with MYD88 L265P-positive DLBCL.

SAN DIEGO—A Toll-like receptor (TLR) antagonist can target B-cell lymphoma cells harboring the MYD88 L265P mutation, preclinical research suggests.
The compound, IMO-8400, decreased the viability of mutated diffuse large B-cell lymphoma (DLBCL) cells and Waldenström’s macroglobulinemia (WM) cells in vitro.
IMO-8400 also decreased tumor growth and prolonged survival in mice with MYD88 L265P-positive DLBCL.
Lakshmi Bhagat, PhD, and colleagues from the Cambridge, Massachusetts-based Idera Pharmaceuticals, Inc.—the company developing IMO-8400—presented these results at the AACR Annual Meeting 2014 (abstract 2570).
The researchers said their data provide additional evidence that the MYD88 L265P mutation results in over-activation of TLR7- and TLR9-mediated signaling, and blocking these TLRs leads to tumor cell death. IMO-8400 is an oligonucleotide-based antagonist of TLRs 7, 8, and 9.
In experiments with OCI‐Ly10 cells (DLBCL cells harboring the MYD88 L265P mutation), IMO-8400 prompted cell death and decreased proliferative cell signaling. But the compound did not produce these effects in SU-DHL-6 cells (DLBCL cells without the MYD88 L265P mutation).
In OCI‐Ly10 cells, IMO-8400 inhibited the IRAK-1, IRAK-4, BTK, STAT-3, Ik-Ba, and NF-κB pathways. The compound did not affect signaling pathways in SU-DHL-6 cells.
IMO-8400 also inhibited tumor growth in a mouse model of MYD88 L265P-positive, activated B-cell-like DLBCL. This inhibition was linked to the suppression of tumor-associated cytokines, including human IL-10, IL-2R, IP-10, and MIG.
Treated mice had significantly longer survival than controls, and the effect was dose-dependent. When IMO-8400 was given at 12.5 mg/kg, the P value was 0.0002. At 25 mg/kg, the P value was less than 0.0002. And at 50 mg/kg, the P value was less than 0.0001.
The researchers also found that IMO‐8400 inhibited cell viability, cytokine production, and signaling pathways in MYD88 L265P-positive WM cells. They observed these effects in the MWCL‐1 cell line and in cells from WM patients.
The team said these results provide a “strong foundation” for accelerating the clinical development of IMO-8400 in patients with B-cell lymphomas harboring the MYD88 L265P mutation.
To that end, Idera has opened enrollment in a phase 1/2 trial of IMO-8400 in WM patients who are refractory to prior therapies. The company has also submitted a protocol to the US Food and Drug Administration to conduct a phase 1/2 trial in patients with MYD88 L265P-positive DLBCL.

SAN DIEGO—A Toll-like receptor (TLR) antagonist can target B-cell lymphoma cells harboring the MYD88 L265P mutation, preclinical research suggests.
The compound, IMO-8400, decreased the viability of mutated diffuse large B-cell lymphoma (DLBCL) cells and Waldenström’s macroglobulinemia (WM) cells in vitro.
IMO-8400 also decreased tumor growth and prolonged survival in mice with MYD88 L265P-positive DLBCL.
Lakshmi Bhagat, PhD, and colleagues from the Cambridge, Massachusetts-based Idera Pharmaceuticals, Inc.—the company developing IMO-8400—presented these results at the AACR Annual Meeting 2014 (abstract 2570).
The researchers said their data provide additional evidence that the MYD88 L265P mutation results in over-activation of TLR7- and TLR9-mediated signaling, and blocking these TLRs leads to tumor cell death. IMO-8400 is an oligonucleotide-based antagonist of TLRs 7, 8, and 9.
In experiments with OCI‐Ly10 cells (DLBCL cells harboring the MYD88 L265P mutation), IMO-8400 prompted cell death and decreased proliferative cell signaling. But the compound did not produce these effects in SU-DHL-6 cells (DLBCL cells without the MYD88 L265P mutation).
In OCI‐Ly10 cells, IMO-8400 inhibited the IRAK-1, IRAK-4, BTK, STAT-3, Ik-Ba, and NF-κB pathways. The compound did not affect signaling pathways in SU-DHL-6 cells.
IMO-8400 also inhibited tumor growth in a mouse model of MYD88 L265P-positive, activated B-cell-like DLBCL. This inhibition was linked to the suppression of tumor-associated cytokines, including human IL-10, IL-2R, IP-10, and MIG.
Treated mice had significantly longer survival than controls, and the effect was dose-dependent. When IMO-8400 was given at 12.5 mg/kg, the P value was 0.0002. At 25 mg/kg, the P value was less than 0.0002. And at 50 mg/kg, the P value was less than 0.0001.
The researchers also found that IMO‐8400 inhibited cell viability, cytokine production, and signaling pathways in MYD88 L265P-positive WM cells. They observed these effects in the MWCL‐1 cell line and in cells from WM patients.
The team said these results provide a “strong foundation” for accelerating the clinical development of IMO-8400 in patients with B-cell lymphomas harboring the MYD88 L265P mutation.
To that end, Idera has opened enrollment in a phase 1/2 trial of IMO-8400 in WM patients who are refractory to prior therapies. The company has also submitted a protocol to the US Food and Drug Administration to conduct a phase 1/2 trial in patients with MYD88 L265P-positive DLBCL.