User login
Master trial seeks to aid drug development for pediatric AML
NEW ORLEANS – Researchers are organizing a master trial in an attempt to improve the treatment of pediatric acute myeloid leukemia (AML).
The Pediatric Acute Leukemia (PedAL) trial is an effort to collect data on all pediatric AML patients. The plan is to use these data to match patients to clinical trials, better understand pediatric AML, and bring new treatments to this population.
E. Anders Kolb, MD, of Nemours Center for Cancer and Blood Disorders in Wilmington, Del., described the initiative at the annual meeting of the American Society of Pediatric Hematology/Oncology.
Dr. Kolb noted that several drugs have been approved to treat adult AML in the last 2 years, but most of them are not approved for use in children.
“What we see in childhood AML is a lot different than what we see in adult AML, and this challenges the paradigm that we have traditionally followed where we use the adult as the 'preclinical model' for pediatric AML,” he said. “I think we are learning more and more that children have a unique disease, unique targets, and need unique therapies.”
The PedAL initiative is an attempt to address these unique needs. PedAL is part of the Leukemia & Lymphoma Society’s Children’s Initiative, and it involves researchers from academic centers and the Children’s Oncology Group.
The PedAL initiative includes preclinical, biomarker, and informatics research, as well as the master clinical trial. The main goal of the master trial is to collect genomic, proteomic, metabolomic, flow cytometry, and clinical data from all children with AML and use these data to match patients to clinical trials.
The PedAL trial will leverage Project:EveryChild, an effort by the Children’s Oncology Group to study every child with cancer. Each child enrolled in this program has an identification number that follows the child through all clinical interventions.
The goal is that Project:EveryChild will capture all pediatric AML patients at the time of diagnosis, although patients can join the project at any time. Then, sequencing, clinical, and other data will be collected from these patients and stored in a data commons.
If patients relapse after standard or other therapies, the GEARBOX algorithm (genomic eligibility algorithm at relapse for better outcomes) can be used to match the patient’s information to clinical trial eligibility criteria and provide a list of appropriate trials.
Dr. Kolb said this process should reduce logistical barriers and get relapsed patients to trials more quickly. Additionally, the data collected through PedAL should help researchers design better trials for pediatric patients with relapsed AML.
“Ultimately, we’ll create the largest data set that will give us a better understanding of all the risks and benefits associated with postrelapse AML,” Dr. Kolb said. “No matter what happens to the patient, no matter where that patient enrolls, we’re going to have the capacity to collect data and present that data to the community for analysis for improved understanding of outcomes.”
Dr. Kolb and his colleagues are already working with researchers in Europe and Japan to make this a global effort and create an international data commons. In addition, the researchers are planning to collaborate with the pharmaceutical industry to unite efforts in pediatric AML drug development.
“We can’t just test drugs in kids because they worked in adults,” Dr. Kolb said. “We really need to maintain the integrity of the science and ask relevant questions in children but do so with the intent to make sure these drugs are licensed for use in kids.”
Dr. Kolb reported having no conflicts of interest. The PedAL trial is sponsored by the Leukemia & Lymphoma Society.
NEW ORLEANS – Researchers are organizing a master trial in an attempt to improve the treatment of pediatric acute myeloid leukemia (AML).
The Pediatric Acute Leukemia (PedAL) trial is an effort to collect data on all pediatric AML patients. The plan is to use these data to match patients to clinical trials, better understand pediatric AML, and bring new treatments to this population.
E. Anders Kolb, MD, of Nemours Center for Cancer and Blood Disorders in Wilmington, Del., described the initiative at the annual meeting of the American Society of Pediatric Hematology/Oncology.
Dr. Kolb noted that several drugs have been approved to treat adult AML in the last 2 years, but most of them are not approved for use in children.
“What we see in childhood AML is a lot different than what we see in adult AML, and this challenges the paradigm that we have traditionally followed where we use the adult as the 'preclinical model' for pediatric AML,” he said. “I think we are learning more and more that children have a unique disease, unique targets, and need unique therapies.”
The PedAL initiative is an attempt to address these unique needs. PedAL is part of the Leukemia & Lymphoma Society’s Children’s Initiative, and it involves researchers from academic centers and the Children’s Oncology Group.
The PedAL initiative includes preclinical, biomarker, and informatics research, as well as the master clinical trial. The main goal of the master trial is to collect genomic, proteomic, metabolomic, flow cytometry, and clinical data from all children with AML and use these data to match patients to clinical trials.
The PedAL trial will leverage Project:EveryChild, an effort by the Children’s Oncology Group to study every child with cancer. Each child enrolled in this program has an identification number that follows the child through all clinical interventions.
The goal is that Project:EveryChild will capture all pediatric AML patients at the time of diagnosis, although patients can join the project at any time. Then, sequencing, clinical, and other data will be collected from these patients and stored in a data commons.
If patients relapse after standard or other therapies, the GEARBOX algorithm (genomic eligibility algorithm at relapse for better outcomes) can be used to match the patient’s information to clinical trial eligibility criteria and provide a list of appropriate trials.
Dr. Kolb said this process should reduce logistical barriers and get relapsed patients to trials more quickly. Additionally, the data collected through PedAL should help researchers design better trials for pediatric patients with relapsed AML.
“Ultimately, we’ll create the largest data set that will give us a better understanding of all the risks and benefits associated with postrelapse AML,” Dr. Kolb said. “No matter what happens to the patient, no matter where that patient enrolls, we’re going to have the capacity to collect data and present that data to the community for analysis for improved understanding of outcomes.”
Dr. Kolb and his colleagues are already working with researchers in Europe and Japan to make this a global effort and create an international data commons. In addition, the researchers are planning to collaborate with the pharmaceutical industry to unite efforts in pediatric AML drug development.
“We can’t just test drugs in kids because they worked in adults,” Dr. Kolb said. “We really need to maintain the integrity of the science and ask relevant questions in children but do so with the intent to make sure these drugs are licensed for use in kids.”
Dr. Kolb reported having no conflicts of interest. The PedAL trial is sponsored by the Leukemia & Lymphoma Society.
NEW ORLEANS – Researchers are organizing a master trial in an attempt to improve the treatment of pediatric acute myeloid leukemia (AML).
The Pediatric Acute Leukemia (PedAL) trial is an effort to collect data on all pediatric AML patients. The plan is to use these data to match patients to clinical trials, better understand pediatric AML, and bring new treatments to this population.
E. Anders Kolb, MD, of Nemours Center for Cancer and Blood Disorders in Wilmington, Del., described the initiative at the annual meeting of the American Society of Pediatric Hematology/Oncology.
Dr. Kolb noted that several drugs have been approved to treat adult AML in the last 2 years, but most of them are not approved for use in children.
“What we see in childhood AML is a lot different than what we see in adult AML, and this challenges the paradigm that we have traditionally followed where we use the adult as the 'preclinical model' for pediatric AML,” he said. “I think we are learning more and more that children have a unique disease, unique targets, and need unique therapies.”
The PedAL initiative is an attempt to address these unique needs. PedAL is part of the Leukemia & Lymphoma Society’s Children’s Initiative, and it involves researchers from academic centers and the Children’s Oncology Group.
The PedAL initiative includes preclinical, biomarker, and informatics research, as well as the master clinical trial. The main goal of the master trial is to collect genomic, proteomic, metabolomic, flow cytometry, and clinical data from all children with AML and use these data to match patients to clinical trials.
The PedAL trial will leverage Project:EveryChild, an effort by the Children’s Oncology Group to study every child with cancer. Each child enrolled in this program has an identification number that follows the child through all clinical interventions.
The goal is that Project:EveryChild will capture all pediatric AML patients at the time of diagnosis, although patients can join the project at any time. Then, sequencing, clinical, and other data will be collected from these patients and stored in a data commons.
If patients relapse after standard or other therapies, the GEARBOX algorithm (genomic eligibility algorithm at relapse for better outcomes) can be used to match the patient’s information to clinical trial eligibility criteria and provide a list of appropriate trials.
Dr. Kolb said this process should reduce logistical barriers and get relapsed patients to trials more quickly. Additionally, the data collected through PedAL should help researchers design better trials for pediatric patients with relapsed AML.
“Ultimately, we’ll create the largest data set that will give us a better understanding of all the risks and benefits associated with postrelapse AML,” Dr. Kolb said. “No matter what happens to the patient, no matter where that patient enrolls, we’re going to have the capacity to collect data and present that data to the community for analysis for improved understanding of outcomes.”
Dr. Kolb and his colleagues are already working with researchers in Europe and Japan to make this a global effort and create an international data commons. In addition, the researchers are planning to collaborate with the pharmaceutical industry to unite efforts in pediatric AML drug development.
“We can’t just test drugs in kids because they worked in adults,” Dr. Kolb said. “We really need to maintain the integrity of the science and ask relevant questions in children but do so with the intent to make sure these drugs are licensed for use in kids.”
Dr. Kolb reported having no conflicts of interest. The PedAL trial is sponsored by the Leukemia & Lymphoma Society.
REPORTING FROM 2019 ASPHO CONFERENCE
Researchers propose new risk groups for NK-AML
NEWPORT BEACH, CALIF. – New research suggests patients with normal karyotype acute myeloid leukemia (NK-AML) can be divided into four risk groups associated with overall survival.
Investigators used machine learning algorithms to study the association between mutations and overall survival in 1,352 patients with NK-AML. The analysis revealed combinations of mutations that could be used to classify NK-AML patients into favorable, intermediate-1, intermediate-2, and unfavorable risk groups.
For example, patients who had NPM1 mutations but wild-type FLT3-ITD and DNMT3A, had a median overall survival of 99.1 months and could be classified as favorable risk. Conversely, patients who had NPM1, FLT3-ITD, and DNMT3A mutations, had a median overall survival of 13.4 months and could be classified as unfavorable risk.
Aziz Nazha, MD, of the Cleveland Clinic, and his colleagues conducted this research and presented the findings at the Acute Leukemia Forum of Hemedicus.
The investigators looked at genomic and clinical data from 1,352 patients with NK-AML. The patients were a median age of 55 years and had a median white blood cell count of 21.3 x 109/L, a median hemoglobin of 9.1 g/dL, and a median platelet count of 61 x 109/L. More than half of patients (57.3%) were male.
The patients were screened for 35 genes that are commonly mutated in AML and other myeloid malignancies. The investigators used machine learning algorithms, including random survival forest and recommender system algorithms, to study the association between mutations and overall survival in an “unbiased” way.
Dr. Nazha said there were a median of three mutations per patient sample, and “there are some competing interests between those mutations to impact the prognosis of the patient.”
The investigators used the mutations and their associations with overall survival to classify patients into the risk groups outlined in the table below.
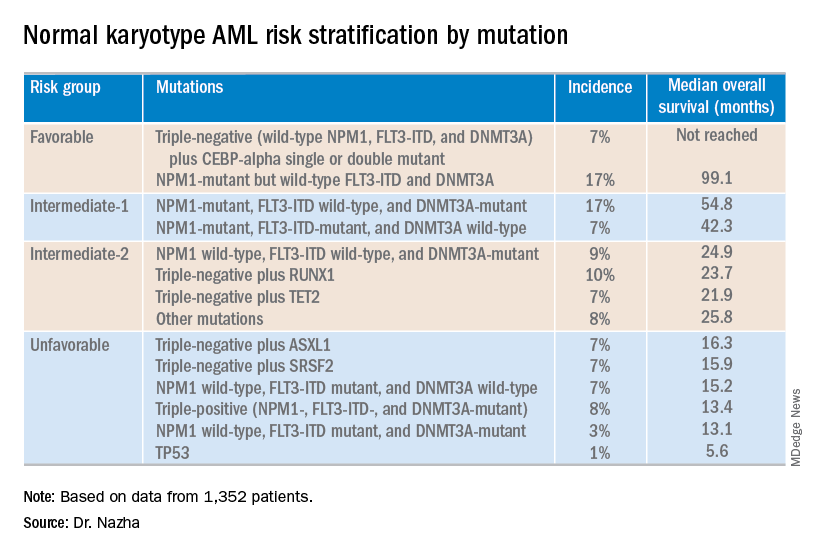
These findings can improve the risk stratification of NK-AML and may aid physicians in making treatment decisions, according to Dr. Nazha and his colleagues. To move this work forward, the investigators are attempting to develop a personalized model that can make predictions specific to an individual patient based on that patient’s mutation information.
Dr. Nazha reported having no financial disclosures relevant to this research. Other investigators reported relationships with the Munich Leukemia Laboratory.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – New research suggests patients with normal karyotype acute myeloid leukemia (NK-AML) can be divided into four risk groups associated with overall survival.
Investigators used machine learning algorithms to study the association between mutations and overall survival in 1,352 patients with NK-AML. The analysis revealed combinations of mutations that could be used to classify NK-AML patients into favorable, intermediate-1, intermediate-2, and unfavorable risk groups.
For example, patients who had NPM1 mutations but wild-type FLT3-ITD and DNMT3A, had a median overall survival of 99.1 months and could be classified as favorable risk. Conversely, patients who had NPM1, FLT3-ITD, and DNMT3A mutations, had a median overall survival of 13.4 months and could be classified as unfavorable risk.
Aziz Nazha, MD, of the Cleveland Clinic, and his colleagues conducted this research and presented the findings at the Acute Leukemia Forum of Hemedicus.
The investigators looked at genomic and clinical data from 1,352 patients with NK-AML. The patients were a median age of 55 years and had a median white blood cell count of 21.3 x 109/L, a median hemoglobin of 9.1 g/dL, and a median platelet count of 61 x 109/L. More than half of patients (57.3%) were male.
The patients were screened for 35 genes that are commonly mutated in AML and other myeloid malignancies. The investigators used machine learning algorithms, including random survival forest and recommender system algorithms, to study the association between mutations and overall survival in an “unbiased” way.
Dr. Nazha said there were a median of three mutations per patient sample, and “there are some competing interests between those mutations to impact the prognosis of the patient.”
The investigators used the mutations and their associations with overall survival to classify patients into the risk groups outlined in the table below.

These findings can improve the risk stratification of NK-AML and may aid physicians in making treatment decisions, according to Dr. Nazha and his colleagues. To move this work forward, the investigators are attempting to develop a personalized model that can make predictions specific to an individual patient based on that patient’s mutation information.
Dr. Nazha reported having no financial disclosures relevant to this research. Other investigators reported relationships with the Munich Leukemia Laboratory.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – New research suggests patients with normal karyotype acute myeloid leukemia (NK-AML) can be divided into four risk groups associated with overall survival.
Investigators used machine learning algorithms to study the association between mutations and overall survival in 1,352 patients with NK-AML. The analysis revealed combinations of mutations that could be used to classify NK-AML patients into favorable, intermediate-1, intermediate-2, and unfavorable risk groups.
For example, patients who had NPM1 mutations but wild-type FLT3-ITD and DNMT3A, had a median overall survival of 99.1 months and could be classified as favorable risk. Conversely, patients who had NPM1, FLT3-ITD, and DNMT3A mutations, had a median overall survival of 13.4 months and could be classified as unfavorable risk.
Aziz Nazha, MD, of the Cleveland Clinic, and his colleagues conducted this research and presented the findings at the Acute Leukemia Forum of Hemedicus.
The investigators looked at genomic and clinical data from 1,352 patients with NK-AML. The patients were a median age of 55 years and had a median white blood cell count of 21.3 x 109/L, a median hemoglobin of 9.1 g/dL, and a median platelet count of 61 x 109/L. More than half of patients (57.3%) were male.
The patients were screened for 35 genes that are commonly mutated in AML and other myeloid malignancies. The investigators used machine learning algorithms, including random survival forest and recommender system algorithms, to study the association between mutations and overall survival in an “unbiased” way.
Dr. Nazha said there were a median of three mutations per patient sample, and “there are some competing interests between those mutations to impact the prognosis of the patient.”
The investigators used the mutations and their associations with overall survival to classify patients into the risk groups outlined in the table below.

These findings can improve the risk stratification of NK-AML and may aid physicians in making treatment decisions, according to Dr. Nazha and his colleagues. To move this work forward, the investigators are attempting to develop a personalized model that can make predictions specific to an individual patient based on that patient’s mutation information.
Dr. Nazha reported having no financial disclosures relevant to this research. Other investigators reported relationships with the Munich Leukemia Laboratory.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
Combo proves most effective in HMA-naive, higher-risk MDS
NEWPORT BEACH, CALIF. – The combination of oral rigosertib and azacitidine is proceeding to a phase 3 trial in patients with myelodysplastic syndromes (MDS), but it isn’t clear if the combination will continue to be developed for acute myeloid leukemia (AML).
In a phase 1/2 trial, oral rigosertib plus azacitidine produced a 90% response rate in higher-risk MDS patients who were naive to hypomethylating agents (HMAs), a 54% response rate in higher-risk MDS patients who had failed HMA therapy, and a 50% response rate in patients with AML.
Genitourinary toxicities were initially a concern in this trial, but researchers found ways to mitigate the risk of these toxicities, according to Richard Woodman, MD, chief medical officer and senior vice president of research and development at Onconova Therapeutics, the company developing rigosertib.
Dr. Woodman and his colleagues presented results from the phase 1/2 trial in two posters at the Acute Leukemia Forum of Hemedicus.
Results in AML
The researchers reported phase 1 results in 17 patients with AML. Eleven patients had AML, according to investigator assessment, and six patients had refractory anemia with excess blasts in transformation, according to French American British criteria, as well as least 20% excess blasts at baseline.
The median age of the patients was 73 years, and 53% were men. Two patients had received no prior therapies, six patients had relapsed disease, and nine were refractory to their last therapy.
Patients received oral rigosertib at escalating doses twice daily on days 1-21 of a 28-day cycle. The recommended phase 2 dose was 840 mg daily (560 mg in the morning and 280 mg in the afternoon), but there were two expansion cohorts in which patients received 1,120 mg daily (560 mg twice a day or 840 mg in the morning and 280 mg in the afternoon). The patients also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
Patients received a median of three treatment cycles. Fifteen of the 17 patients (88%) discontinued treatment, most because of progressive disease (n = 5), toxicity (n = 4), or death (n = 3).
Twelve patients were evaluable for response, and six (50%) responded. One patient achieved a morphologic complete remission (CR), three achieved a morphologic leukemia-free state, and two had a partial response.
The most common treatment-emergent adverse events (TEAEs) were fatigue (53%), diarrhea (53%), nausea (53%), constipation (47%), back pain (41%), pyrexia (41%), and pneumonia (35%). Grade 3 or higher TEAEs included pneumonia (35%) and anemia (24%).
These results haven’t provided a clear way forward for oral rigosertib and azacitidine in AML. Dr. Woodman said the researchers will have to review past studies and evaluate how AML patients (with at least 20% blasts) have responded to intravenous rigosertib, consult experts in the field, and then decide how they will move forward with oral rigosertib and azacitidine in AML.
Results in MDS
Dr. Woodman and his colleagues presented data on 74 patients with higher-risk MDS. The median age was 69 years, and 59% were men. Most patients were high risk (n = 23) or very high risk (n = 33), according to the Revised International Prognostic Scoring System.
The patients received oral rigosertib at a dose of 840 mg/day or higher on days 1-21 of a 28-day cycle. They also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
The median duration of treatment was 7.8 months in patients who were HMA naive and 4.9 months in patients who failed HMA therapy. The most common reasons for treatment discontinuation in the HMA-naive patients were toxicity (n = 8), progression (n = 7), and patient request (n = 7). The most common reasons for discontinuation in patients who had failed HMA therapy were progression (n = 12), toxicity (n = 5), and investigator decision (n = 4).
In total, 55 patients were evaluable for response, 26 who had failed HMA therapy and 29 who were HMA naive.
“The best responses, not surprisingly, were in patients that were HMA naive,” Dr. Woodman said.
In the HMA-naive patients, the overall response rate was 90%. Ten patients had a CR, five had a marrow CR with hematologic improvement, three had hematologic improvement alone, eight had a marrow CR alone, and three patients had stable disease. None of the patients progressed.
In the patients who had failed HMA therapy, the overall response rate was 54%. One patient achieved a CR, one had a partial response, five had a marrow CR with hematologic improvement, two had hematologic improvement alone, five had a marrow CR alone, seven had stable disease, and five progressed.
The median duration of response was 10.8 months in patients who failed HMA therapy and 12.2 months in the HMA-naive patients.
The most common TEAEs in the entire MDS cohort were hematuria (45%), constipation (43%), diarrhea (42%), fatigue (42%), dysuria (38%), pyrexia (36%), nausea (35%), neutropenia (31%), and thrombocytopenia (30%).
Grade 3 or higher TEAEs were neutropenia (27%), thrombocytopenia (26%), hematuria (9%), dysuria (9%), diarrhea (5%), fatigue (4%), and pyrexia (1%).
Dr. Woodman said patients who were most likely to be at risk for genitourinary toxicities (hematuria and dysuria) were those who weren’t well hydrated, took rigosertib at night, and didn’t void their bladders before bedtime. He said the researchers’ hypothesis is that there is some local bladder irritation in that setting.
However, the researchers found ways to mitigate the risk of genitourinary toxicities, including:
- Requiring the second dose of rigosertib to be taken in the afternoon rather than evening (about 3 p.m.).
- Asking patients to consume at least 2 liters of fluid per day.
- Having patients empty their bladders before bedtime.
- Assessing urine pH roughly 2 hours after the morning dose of rigosertib and prescribing sodium bicarbonate if the pH is less than 7.5.
Dr. Woodman said the phase 2 results in MDS patients have prompted the development of a phase 3 trial in which researchers will compare oral rigosertib plus azacitidine to azacitidine plus placebo.
Dr. Woodman is employed by Onconova Therapeutics, which sponsored the phase 1/2 trial. The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The combination of oral rigosertib and azacitidine is proceeding to a phase 3 trial in patients with myelodysplastic syndromes (MDS), but it isn’t clear if the combination will continue to be developed for acute myeloid leukemia (AML).
In a phase 1/2 trial, oral rigosertib plus azacitidine produced a 90% response rate in higher-risk MDS patients who were naive to hypomethylating agents (HMAs), a 54% response rate in higher-risk MDS patients who had failed HMA therapy, and a 50% response rate in patients with AML.
Genitourinary toxicities were initially a concern in this trial, but researchers found ways to mitigate the risk of these toxicities, according to Richard Woodman, MD, chief medical officer and senior vice president of research and development at Onconova Therapeutics, the company developing rigosertib.
Dr. Woodman and his colleagues presented results from the phase 1/2 trial in two posters at the Acute Leukemia Forum of Hemedicus.
Results in AML
The researchers reported phase 1 results in 17 patients with AML. Eleven patients had AML, according to investigator assessment, and six patients had refractory anemia with excess blasts in transformation, according to French American British criteria, as well as least 20% excess blasts at baseline.
The median age of the patients was 73 years, and 53% were men. Two patients had received no prior therapies, six patients had relapsed disease, and nine were refractory to their last therapy.
Patients received oral rigosertib at escalating doses twice daily on days 1-21 of a 28-day cycle. The recommended phase 2 dose was 840 mg daily (560 mg in the morning and 280 mg in the afternoon), but there were two expansion cohorts in which patients received 1,120 mg daily (560 mg twice a day or 840 mg in the morning and 280 mg in the afternoon). The patients also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
Patients received a median of three treatment cycles. Fifteen of the 17 patients (88%) discontinued treatment, most because of progressive disease (n = 5), toxicity (n = 4), or death (n = 3).
Twelve patients were evaluable for response, and six (50%) responded. One patient achieved a morphologic complete remission (CR), three achieved a morphologic leukemia-free state, and two had a partial response.
The most common treatment-emergent adverse events (TEAEs) were fatigue (53%), diarrhea (53%), nausea (53%), constipation (47%), back pain (41%), pyrexia (41%), and pneumonia (35%). Grade 3 or higher TEAEs included pneumonia (35%) and anemia (24%).
These results haven’t provided a clear way forward for oral rigosertib and azacitidine in AML. Dr. Woodman said the researchers will have to review past studies and evaluate how AML patients (with at least 20% blasts) have responded to intravenous rigosertib, consult experts in the field, and then decide how they will move forward with oral rigosertib and azacitidine in AML.
Results in MDS
Dr. Woodman and his colleagues presented data on 74 patients with higher-risk MDS. The median age was 69 years, and 59% were men. Most patients were high risk (n = 23) or very high risk (n = 33), according to the Revised International Prognostic Scoring System.
The patients received oral rigosertib at a dose of 840 mg/day or higher on days 1-21 of a 28-day cycle. They also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
The median duration of treatment was 7.8 months in patients who were HMA naive and 4.9 months in patients who failed HMA therapy. The most common reasons for treatment discontinuation in the HMA-naive patients were toxicity (n = 8), progression (n = 7), and patient request (n = 7). The most common reasons for discontinuation in patients who had failed HMA therapy were progression (n = 12), toxicity (n = 5), and investigator decision (n = 4).
In total, 55 patients were evaluable for response, 26 who had failed HMA therapy and 29 who were HMA naive.
“The best responses, not surprisingly, were in patients that were HMA naive,” Dr. Woodman said.
In the HMA-naive patients, the overall response rate was 90%. Ten patients had a CR, five had a marrow CR with hematologic improvement, three had hematologic improvement alone, eight had a marrow CR alone, and three patients had stable disease. None of the patients progressed.
In the patients who had failed HMA therapy, the overall response rate was 54%. One patient achieved a CR, one had a partial response, five had a marrow CR with hematologic improvement, two had hematologic improvement alone, five had a marrow CR alone, seven had stable disease, and five progressed.
The median duration of response was 10.8 months in patients who failed HMA therapy and 12.2 months in the HMA-naive patients.
The most common TEAEs in the entire MDS cohort were hematuria (45%), constipation (43%), diarrhea (42%), fatigue (42%), dysuria (38%), pyrexia (36%), nausea (35%), neutropenia (31%), and thrombocytopenia (30%).
Grade 3 or higher TEAEs were neutropenia (27%), thrombocytopenia (26%), hematuria (9%), dysuria (9%), diarrhea (5%), fatigue (4%), and pyrexia (1%).
Dr. Woodman said patients who were most likely to be at risk for genitourinary toxicities (hematuria and dysuria) were those who weren’t well hydrated, took rigosertib at night, and didn’t void their bladders before bedtime. He said the researchers’ hypothesis is that there is some local bladder irritation in that setting.
However, the researchers found ways to mitigate the risk of genitourinary toxicities, including:
- Requiring the second dose of rigosertib to be taken in the afternoon rather than evening (about 3 p.m.).
- Asking patients to consume at least 2 liters of fluid per day.
- Having patients empty their bladders before bedtime.
- Assessing urine pH roughly 2 hours after the morning dose of rigosertib and prescribing sodium bicarbonate if the pH is less than 7.5.
Dr. Woodman said the phase 2 results in MDS patients have prompted the development of a phase 3 trial in which researchers will compare oral rigosertib plus azacitidine to azacitidine plus placebo.
Dr. Woodman is employed by Onconova Therapeutics, which sponsored the phase 1/2 trial. The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The combination of oral rigosertib and azacitidine is proceeding to a phase 3 trial in patients with myelodysplastic syndromes (MDS), but it isn’t clear if the combination will continue to be developed for acute myeloid leukemia (AML).
In a phase 1/2 trial, oral rigosertib plus azacitidine produced a 90% response rate in higher-risk MDS patients who were naive to hypomethylating agents (HMAs), a 54% response rate in higher-risk MDS patients who had failed HMA therapy, and a 50% response rate in patients with AML.
Genitourinary toxicities were initially a concern in this trial, but researchers found ways to mitigate the risk of these toxicities, according to Richard Woodman, MD, chief medical officer and senior vice president of research and development at Onconova Therapeutics, the company developing rigosertib.
Dr. Woodman and his colleagues presented results from the phase 1/2 trial in two posters at the Acute Leukemia Forum of Hemedicus.
Results in AML
The researchers reported phase 1 results in 17 patients with AML. Eleven patients had AML, according to investigator assessment, and six patients had refractory anemia with excess blasts in transformation, according to French American British criteria, as well as least 20% excess blasts at baseline.
The median age of the patients was 73 years, and 53% were men. Two patients had received no prior therapies, six patients had relapsed disease, and nine were refractory to their last therapy.
Patients received oral rigosertib at escalating doses twice daily on days 1-21 of a 28-day cycle. The recommended phase 2 dose was 840 mg daily (560 mg in the morning and 280 mg in the afternoon), but there were two expansion cohorts in which patients received 1,120 mg daily (560 mg twice a day or 840 mg in the morning and 280 mg in the afternoon). The patients also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
Patients received a median of three treatment cycles. Fifteen of the 17 patients (88%) discontinued treatment, most because of progressive disease (n = 5), toxicity (n = 4), or death (n = 3).
Twelve patients were evaluable for response, and six (50%) responded. One patient achieved a morphologic complete remission (CR), three achieved a morphologic leukemia-free state, and two had a partial response.
The most common treatment-emergent adverse events (TEAEs) were fatigue (53%), diarrhea (53%), nausea (53%), constipation (47%), back pain (41%), pyrexia (41%), and pneumonia (35%). Grade 3 or higher TEAEs included pneumonia (35%) and anemia (24%).
These results haven’t provided a clear way forward for oral rigosertib and azacitidine in AML. Dr. Woodman said the researchers will have to review past studies and evaluate how AML patients (with at least 20% blasts) have responded to intravenous rigosertib, consult experts in the field, and then decide how they will move forward with oral rigosertib and azacitidine in AML.
Results in MDS
Dr. Woodman and his colleagues presented data on 74 patients with higher-risk MDS. The median age was 69 years, and 59% were men. Most patients were high risk (n = 23) or very high risk (n = 33), according to the Revised International Prognostic Scoring System.
The patients received oral rigosertib at a dose of 840 mg/day or higher on days 1-21 of a 28-day cycle. They also received azacitidine at 75 mg/m2 per day subcutaneously or intravenously for 7 days starting on day 8.
The median duration of treatment was 7.8 months in patients who were HMA naive and 4.9 months in patients who failed HMA therapy. The most common reasons for treatment discontinuation in the HMA-naive patients were toxicity (n = 8), progression (n = 7), and patient request (n = 7). The most common reasons for discontinuation in patients who had failed HMA therapy were progression (n = 12), toxicity (n = 5), and investigator decision (n = 4).
In total, 55 patients were evaluable for response, 26 who had failed HMA therapy and 29 who were HMA naive.
“The best responses, not surprisingly, were in patients that were HMA naive,” Dr. Woodman said.
In the HMA-naive patients, the overall response rate was 90%. Ten patients had a CR, five had a marrow CR with hematologic improvement, three had hematologic improvement alone, eight had a marrow CR alone, and three patients had stable disease. None of the patients progressed.
In the patients who had failed HMA therapy, the overall response rate was 54%. One patient achieved a CR, one had a partial response, five had a marrow CR with hematologic improvement, two had hematologic improvement alone, five had a marrow CR alone, seven had stable disease, and five progressed.
The median duration of response was 10.8 months in patients who failed HMA therapy and 12.2 months in the HMA-naive patients.
The most common TEAEs in the entire MDS cohort were hematuria (45%), constipation (43%), diarrhea (42%), fatigue (42%), dysuria (38%), pyrexia (36%), nausea (35%), neutropenia (31%), and thrombocytopenia (30%).
Grade 3 or higher TEAEs were neutropenia (27%), thrombocytopenia (26%), hematuria (9%), dysuria (9%), diarrhea (5%), fatigue (4%), and pyrexia (1%).
Dr. Woodman said patients who were most likely to be at risk for genitourinary toxicities (hematuria and dysuria) were those who weren’t well hydrated, took rigosertib at night, and didn’t void their bladders before bedtime. He said the researchers’ hypothesis is that there is some local bladder irritation in that setting.
However, the researchers found ways to mitigate the risk of genitourinary toxicities, including:
- Requiring the second dose of rigosertib to be taken in the afternoon rather than evening (about 3 p.m.).
- Asking patients to consume at least 2 liters of fluid per day.
- Having patients empty their bladders before bedtime.
- Assessing urine pH roughly 2 hours after the morning dose of rigosertib and prescribing sodium bicarbonate if the pH is less than 7.5.
Dr. Woodman said the phase 2 results in MDS patients have prompted the development of a phase 3 trial in which researchers will compare oral rigosertib plus azacitidine to azacitidine plus placebo.
Dr. Woodman is employed by Onconova Therapeutics, which sponsored the phase 1/2 trial. The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
FDA approves ivosidenib frontline for certain AML patients
The Food and Drug Administration has approved ivosidenib (Tibsovo) for newly diagnosed acute myeloid leukemia (AML) with a susceptible IDH1 mutation in patients who are at least 75 years old or have comorbidities preventing the use of intensive induction chemotherapy.
In July 2018, the FDA approved ivosidenib for adults with relapsed or refractory AML with a susceptible IDH1 mutation.
The latest approval was based on results from an open-label, single-arm, multicenter trial of patients with newly diagnosed AML with an IDH1 mutation. Patients were treated with 500 mg ivosidenib daily until disease progression, development of unacceptable toxicity, or hematopoietic stem cell transplantation; the median age of the 28 patients treated with ivosidenib was 77 years.
Of the 28 patients treated, 12 achieved complete remission or complete remission with partial hematologic recovery; 7 of the 17 transfusion-dependent patients achieved transfusion independence for at least 8 weeks.
The most common adverse events were diarrhea, fatigue, edema, decreased appetite, leukocytosis, nausea, arthralgia, abdominal pain, dyspnea, differentiation syndrome, and myalgia. The drug’s prescribing information includes a boxed warning on the risk of differentiation syndrome.
“The recommended ivosidenib dose is 500 mg orally once daily with or without food until disease progression or unacceptable toxicity. For patients without disease progression or unacceptable toxicity, treatment is recommended for a minimum of 6 months to allow time for clinical response,” the FDA noted.
Find the full press release on the FDA website.
The Food and Drug Administration has approved ivosidenib (Tibsovo) for newly diagnosed acute myeloid leukemia (AML) with a susceptible IDH1 mutation in patients who are at least 75 years old or have comorbidities preventing the use of intensive induction chemotherapy.
In July 2018, the FDA approved ivosidenib for adults with relapsed or refractory AML with a susceptible IDH1 mutation.
The latest approval was based on results from an open-label, single-arm, multicenter trial of patients with newly diagnosed AML with an IDH1 mutation. Patients were treated with 500 mg ivosidenib daily until disease progression, development of unacceptable toxicity, or hematopoietic stem cell transplantation; the median age of the 28 patients treated with ivosidenib was 77 years.
Of the 28 patients treated, 12 achieved complete remission or complete remission with partial hematologic recovery; 7 of the 17 transfusion-dependent patients achieved transfusion independence for at least 8 weeks.
The most common adverse events were diarrhea, fatigue, edema, decreased appetite, leukocytosis, nausea, arthralgia, abdominal pain, dyspnea, differentiation syndrome, and myalgia. The drug’s prescribing information includes a boxed warning on the risk of differentiation syndrome.
“The recommended ivosidenib dose is 500 mg orally once daily with or without food until disease progression or unacceptable toxicity. For patients without disease progression or unacceptable toxicity, treatment is recommended for a minimum of 6 months to allow time for clinical response,” the FDA noted.
Find the full press release on the FDA website.
The Food and Drug Administration has approved ivosidenib (Tibsovo) for newly diagnosed acute myeloid leukemia (AML) with a susceptible IDH1 mutation in patients who are at least 75 years old or have comorbidities preventing the use of intensive induction chemotherapy.
In July 2018, the FDA approved ivosidenib for adults with relapsed or refractory AML with a susceptible IDH1 mutation.
The latest approval was based on results from an open-label, single-arm, multicenter trial of patients with newly diagnosed AML with an IDH1 mutation. Patients were treated with 500 mg ivosidenib daily until disease progression, development of unacceptable toxicity, or hematopoietic stem cell transplantation; the median age of the 28 patients treated with ivosidenib was 77 years.
Of the 28 patients treated, 12 achieved complete remission or complete remission with partial hematologic recovery; 7 of the 17 transfusion-dependent patients achieved transfusion independence for at least 8 weeks.
The most common adverse events were diarrhea, fatigue, edema, decreased appetite, leukocytosis, nausea, arthralgia, abdominal pain, dyspnea, differentiation syndrome, and myalgia. The drug’s prescribing information includes a boxed warning on the risk of differentiation syndrome.
“The recommended ivosidenib dose is 500 mg orally once daily with or without food until disease progression or unacceptable toxicity. For patients without disease progression or unacceptable toxicity, treatment is recommended for a minimum of 6 months to allow time for clinical response,” the FDA noted.
Find the full press release on the FDA website.
Sorafenib plus GCLAM held safe in AML, MDS phase-1 study
NEWPORT BEACH, CALIF. – A five-drug regimen was deemed safe in patients with newly diagnosed acute myeloid leukemia (AML) or high-risk myelodysplastic syndromes (MDS), and it appeared to be effective regardless of patients’ FLT3 status.
Researchers tested this regimen – sorafenib plus granulocyte colony–stimulating factor (G-CSF), cladribine, high-dose cytarabine, and mitoxantrone (GCLAM) – in a phase 1 trial.
Kelsey-Leigh Garcia, a clinical research coordinator at Seattle Cancer Care Alliance, and her colleagues presented the results at the Acute Leukemia Forum of Hemedicus.
“The background for doing this study was our institutional results of GCLAM [Leukemia. 2018 Nov;32(11):2352-62] that showed a higher minimal residual disease–negative complete response rate than 7+3 [cytarabine continuously for 7 days, along with short infusions of an anthracycline on each of the first 3 days] and an international study by Röllig that showed the addition of sorafenib to 7+3 increased event-free survival versus [7+3 and] placebo [Lancet Oncol. 2015 Dec;16(16):1691-9],” Ms. Garcia said.
“GCLAM is the standard backbone at our institution, and we wanted to ask the question, ‘If we add sorafenib, can this improve upon the results of GCLAM?’ ” said Anna Halpern, MD, a hematologist-oncologist at the University of Washington, Seattle and principal investigator of the phase 1 trial.
The trial (NCT02728050) included 47 patients, 39 with AML and 8 with MDS. Patients were aged 60 years or younger and had a median age of 48. They had a median treatment-related mortality score of 1.76 (range, 0.19-12.26). A total of 11 patients (23%) had FLT3-ITD, and 4 (9%) had FLT3-TKD.
Treatment and toxicity
For induction, patients received G-CSF at 5 mcg/kg on days 0-5, cladribine at 5 mg/m2 on days 1-5, and cytarabine at 2 g/m2 on days 1-5. Mitoxantrone was given at 10 mg/m2, 12 mg/m2, 15 mg/m2, or 18 mg/m2 on days 1-3. Sorafenib was given at 200 mg twice daily, 400 mg in the morning and 200 mg in the afternoon, or 400 mg b.i.d. on days 10-19.
For consolidation, patients could receive up to four cycles of G-CSF, cladribine, and cytarabine plus sorafenib on days 8-27. Patients who did not proceed to transplant could receive 12 months of sorafenib as maintenance therapy.
There were four dose-limiting toxicities.
- Grade 4 intracranial hemorrhage with mitoxantrone at 12 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 prolonged count recovery with mitoxantrone at 15 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 sepsis, Sweet syndrome, and Bell’s palsy with mitoxantrone at 18 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 3 cardiomyopathy and acute pericarditis with mitoxantrone at 18 mg/m2 and sorafenib at 400 mg b.i.d.
However, these toxicities did not define the maximum-tolerated dose. Therefore, the recommended phase 2 dose of mitoxantrone is 18 mg/m2, and the recommended phase 2 dose of sorafenib is 400 mg b.i.d.
There were no grade 5 treatment-related adverse events. Grade 3 events included febrile neutropenia (90%), maculopapular rash (20%), infections (10%), hand-foot syndrome (2%), and diarrhea (1%). Grade 4 events included sepsis, intracranial hemorrhage, and oral mucositis (all 1%).
Response and survival
Among the 46 evaluable patients, 83% achieved a complete response, 78% had a minimal residual disease–negative complete response, and 4% had a minimal residual disease–negative complete response with incomplete count recovery. A morphological leukemia-free state was achieved by 4% of patients, and 8% had resistant disease.
Fifty-nine percent of patients went on to transplant. The median overall survival had not been reached at a median follow-up of 10 months.
The researchers compared outcomes in this trial with outcomes in a cohort of patients who had received GCLAM alone, and there were no significant differences in overall survival or event-free survival.
“The trial wasn’t powered, necessarily, for efficacy, but we compared these results to our historical cohort of medically matched and age-matched patients treated with GCLAM alone and, so far, found no differences in survival between the two groups,” Dr. Halpern said.
She noted, however, that follow-up was short in the sorafenib trial, and it included patients treated with all dose levels of sorafenib and mitoxantrone.
A phase 2 study of sorafenib plus GCLAM in newly diagnosed AML or high-risk MDS is now underway.
Dr. Halpern and Ms. Garcia reported that they had no conflicts of interest. The phase 1 trial was sponsored by the University of Washington in collaboration with the National Cancer Institute, and funding was provided by Bayer.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – A five-drug regimen was deemed safe in patients with newly diagnosed acute myeloid leukemia (AML) or high-risk myelodysplastic syndromes (MDS), and it appeared to be effective regardless of patients’ FLT3 status.
Researchers tested this regimen – sorafenib plus granulocyte colony–stimulating factor (G-CSF), cladribine, high-dose cytarabine, and mitoxantrone (GCLAM) – in a phase 1 trial.
Kelsey-Leigh Garcia, a clinical research coordinator at Seattle Cancer Care Alliance, and her colleagues presented the results at the Acute Leukemia Forum of Hemedicus.
“The background for doing this study was our institutional results of GCLAM [Leukemia. 2018 Nov;32(11):2352-62] that showed a higher minimal residual disease–negative complete response rate than 7+3 [cytarabine continuously for 7 days, along with short infusions of an anthracycline on each of the first 3 days] and an international study by Röllig that showed the addition of sorafenib to 7+3 increased event-free survival versus [7+3 and] placebo [Lancet Oncol. 2015 Dec;16(16):1691-9],” Ms. Garcia said.
“GCLAM is the standard backbone at our institution, and we wanted to ask the question, ‘If we add sorafenib, can this improve upon the results of GCLAM?’ ” said Anna Halpern, MD, a hematologist-oncologist at the University of Washington, Seattle and principal investigator of the phase 1 trial.
The trial (NCT02728050) included 47 patients, 39 with AML and 8 with MDS. Patients were aged 60 years or younger and had a median age of 48. They had a median treatment-related mortality score of 1.76 (range, 0.19-12.26). A total of 11 patients (23%) had FLT3-ITD, and 4 (9%) had FLT3-TKD.
Treatment and toxicity
For induction, patients received G-CSF at 5 mcg/kg on days 0-5, cladribine at 5 mg/m2 on days 1-5, and cytarabine at 2 g/m2 on days 1-5. Mitoxantrone was given at 10 mg/m2, 12 mg/m2, 15 mg/m2, or 18 mg/m2 on days 1-3. Sorafenib was given at 200 mg twice daily, 400 mg in the morning and 200 mg in the afternoon, or 400 mg b.i.d. on days 10-19.
For consolidation, patients could receive up to four cycles of G-CSF, cladribine, and cytarabine plus sorafenib on days 8-27. Patients who did not proceed to transplant could receive 12 months of sorafenib as maintenance therapy.
There were four dose-limiting toxicities.
- Grade 4 intracranial hemorrhage with mitoxantrone at 12 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 prolonged count recovery with mitoxantrone at 15 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 sepsis, Sweet syndrome, and Bell’s palsy with mitoxantrone at 18 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 3 cardiomyopathy and acute pericarditis with mitoxantrone at 18 mg/m2 and sorafenib at 400 mg b.i.d.
However, these toxicities did not define the maximum-tolerated dose. Therefore, the recommended phase 2 dose of mitoxantrone is 18 mg/m2, and the recommended phase 2 dose of sorafenib is 400 mg b.i.d.
There were no grade 5 treatment-related adverse events. Grade 3 events included febrile neutropenia (90%), maculopapular rash (20%), infections (10%), hand-foot syndrome (2%), and diarrhea (1%). Grade 4 events included sepsis, intracranial hemorrhage, and oral mucositis (all 1%).
Response and survival
Among the 46 evaluable patients, 83% achieved a complete response, 78% had a minimal residual disease–negative complete response, and 4% had a minimal residual disease–negative complete response with incomplete count recovery. A morphological leukemia-free state was achieved by 4% of patients, and 8% had resistant disease.
Fifty-nine percent of patients went on to transplant. The median overall survival had not been reached at a median follow-up of 10 months.
The researchers compared outcomes in this trial with outcomes in a cohort of patients who had received GCLAM alone, and there were no significant differences in overall survival or event-free survival.
“The trial wasn’t powered, necessarily, for efficacy, but we compared these results to our historical cohort of medically matched and age-matched patients treated with GCLAM alone and, so far, found no differences in survival between the two groups,” Dr. Halpern said.
She noted, however, that follow-up was short in the sorafenib trial, and it included patients treated with all dose levels of sorafenib and mitoxantrone.
A phase 2 study of sorafenib plus GCLAM in newly diagnosed AML or high-risk MDS is now underway.
Dr. Halpern and Ms. Garcia reported that they had no conflicts of interest. The phase 1 trial was sponsored by the University of Washington in collaboration with the National Cancer Institute, and funding was provided by Bayer.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – A five-drug regimen was deemed safe in patients with newly diagnosed acute myeloid leukemia (AML) or high-risk myelodysplastic syndromes (MDS), and it appeared to be effective regardless of patients’ FLT3 status.
Researchers tested this regimen – sorafenib plus granulocyte colony–stimulating factor (G-CSF), cladribine, high-dose cytarabine, and mitoxantrone (GCLAM) – in a phase 1 trial.
Kelsey-Leigh Garcia, a clinical research coordinator at Seattle Cancer Care Alliance, and her colleagues presented the results at the Acute Leukemia Forum of Hemedicus.
“The background for doing this study was our institutional results of GCLAM [Leukemia. 2018 Nov;32(11):2352-62] that showed a higher minimal residual disease–negative complete response rate than 7+3 [cytarabine continuously for 7 days, along with short infusions of an anthracycline on each of the first 3 days] and an international study by Röllig that showed the addition of sorafenib to 7+3 increased event-free survival versus [7+3 and] placebo [Lancet Oncol. 2015 Dec;16(16):1691-9],” Ms. Garcia said.
“GCLAM is the standard backbone at our institution, and we wanted to ask the question, ‘If we add sorafenib, can this improve upon the results of GCLAM?’ ” said Anna Halpern, MD, a hematologist-oncologist at the University of Washington, Seattle and principal investigator of the phase 1 trial.
The trial (NCT02728050) included 47 patients, 39 with AML and 8 with MDS. Patients were aged 60 years or younger and had a median age of 48. They had a median treatment-related mortality score of 1.76 (range, 0.19-12.26). A total of 11 patients (23%) had FLT3-ITD, and 4 (9%) had FLT3-TKD.
Treatment and toxicity
For induction, patients received G-CSF at 5 mcg/kg on days 0-5, cladribine at 5 mg/m2 on days 1-5, and cytarabine at 2 g/m2 on days 1-5. Mitoxantrone was given at 10 mg/m2, 12 mg/m2, 15 mg/m2, or 18 mg/m2 on days 1-3. Sorafenib was given at 200 mg twice daily, 400 mg in the morning and 200 mg in the afternoon, or 400 mg b.i.d. on days 10-19.
For consolidation, patients could receive up to four cycles of G-CSF, cladribine, and cytarabine plus sorafenib on days 8-27. Patients who did not proceed to transplant could receive 12 months of sorafenib as maintenance therapy.
There were four dose-limiting toxicities.
- Grade 4 intracranial hemorrhage with mitoxantrone at 12 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 prolonged count recovery with mitoxantrone at 15 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 4 sepsis, Sweet syndrome, and Bell’s palsy with mitoxantrone at 18 mg/m2 and sorafenib at 200 mg b.i.d.
- Grade 3 cardiomyopathy and acute pericarditis with mitoxantrone at 18 mg/m2 and sorafenib at 400 mg b.i.d.
However, these toxicities did not define the maximum-tolerated dose. Therefore, the recommended phase 2 dose of mitoxantrone is 18 mg/m2, and the recommended phase 2 dose of sorafenib is 400 mg b.i.d.
There were no grade 5 treatment-related adverse events. Grade 3 events included febrile neutropenia (90%), maculopapular rash (20%), infections (10%), hand-foot syndrome (2%), and diarrhea (1%). Grade 4 events included sepsis, intracranial hemorrhage, and oral mucositis (all 1%).
Response and survival
Among the 46 evaluable patients, 83% achieved a complete response, 78% had a minimal residual disease–negative complete response, and 4% had a minimal residual disease–negative complete response with incomplete count recovery. A morphological leukemia-free state was achieved by 4% of patients, and 8% had resistant disease.
Fifty-nine percent of patients went on to transplant. The median overall survival had not been reached at a median follow-up of 10 months.
The researchers compared outcomes in this trial with outcomes in a cohort of patients who had received GCLAM alone, and there were no significant differences in overall survival or event-free survival.
“The trial wasn’t powered, necessarily, for efficacy, but we compared these results to our historical cohort of medically matched and age-matched patients treated with GCLAM alone and, so far, found no differences in survival between the two groups,” Dr. Halpern said.
She noted, however, that follow-up was short in the sorafenib trial, and it included patients treated with all dose levels of sorafenib and mitoxantrone.
A phase 2 study of sorafenib plus GCLAM in newly diagnosed AML or high-risk MDS is now underway.
Dr. Halpern and Ms. Garcia reported that they had no conflicts of interest. The phase 1 trial was sponsored by the University of Washington in collaboration with the National Cancer Institute, and funding was provided by Bayer.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
Study highlights lack of data on transgender leukemia patients
NEWPORT BEACH, CALIF. – Researchers have shown they can identify transgender leukemia patients by detecting gender-karyotype mismatches, but some transgender patients may be overlooked with this method.
The researchers’ work also highlights how little we know about transgender patients with leukemia and other cancers.
Alison Alpert, MD, of the University of Rochester (N.Y.) Medical Center, and her colleagues conducted this research and presented their findings in a poster at the Acute Leukemia Forum of Hemedicus.
“There’s almost no data about transgender people with cancer ... in terms of prevalence or anything else,” Dr. Alpert noted. “And because we don’t know which patients with cancer are transgender, we can’t begin to answer any of the other big questions for patients.”
Specifically, it’s unclear what kinds of cancer transgender patients have, if there are health disparities among transgender patients, if it is safe to continue hormone therapy during cancer treatment, and if it is possible to do transition-related surgeries in the context of cancer care.
With this in mind, Dr. Alpert and her colleagues set out to identify transgender patients by detecting gender-karyotype mismatches. The team analyzed data on patients with acute myeloid leukemia (AML) or myelodysplastic syndromes enrolled in five Southwest Oncology Group (SWOG) trials.
Of the 1,748 patients analyzed, six (0.3%) had a gender-karyotype mismatch. Five patients had a 46,XY karyotype and identified as female, and one patient had a 46,XX karyotype and identified as male.
“Some transgender patients have their gender identity accurately reflected in the electronic medical record, [but] some transgender patients probably don’t,” Dr. Alpert noted. “So we identified some, but probably not all, and probably not even most, transgender patients with leukemia in this cohort.”
All six of the transgender patients identified had AML, and all were white. They ranged in age from 18 to 57 years. Four patients had achieved a complete response to therapy, and two had refractory disease.
Four patients, including one who was refractory, were still alive at last follow-up. The remaining two patients, including one who had achieved a complete response, had died.
The transgender patients identified in this analysis represent a very small percentage of the population studied, Dr. Alpert noted. Therefore, the researchers could not draw any conclusions about transgender patients with AML.
“Mostly, what we did was, we pointed out how little information we have,” Dr. Alpert said. “Oncologists don’t routinely collect gender identity information, and this information doesn’t exist in cooperative group databases either.”
“But going forward, what probably really needs to happen is that oncologists need to ask their patients whether they are transgender or not. And then, ideally, consent forms for large cooperative groups like SWOG would include gender identity data, and then we would be able to answer some of our other questions and better counsel our patients.”
Dr. Alpert and her colleagues are hoping to gain insights regarding transgender patients with lymphoma as well. The researchers are analyzing the lymphoma database at the University of Rochester Medical Center, which includes about 2,200 patients.
The team is attempting to identify transgender lymphoma patients using gender-karyotype mismatch as well as other methods, including assessing patients’ medication and surgical histories, determining whether patients have any aliases, and looking for the word “transgender” in patient charts.
“Given that the country is finally starting to talk about transgender patients, their health disparities, and their needs and experiences, it’s really time that we start collecting this data,” Dr. Alpert said.
“[I]f we are able to start to collect this data, it can help us build relationships with our patients, improve their care and outcomes, and, hopefully, be able to better counsel them about hormones and surgery.”
Dr. Alpert and her colleagues did not disclose any conflicts of interest.
The Acute Leukemia Forum is organized by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – Researchers have shown they can identify transgender leukemia patients by detecting gender-karyotype mismatches, but some transgender patients may be overlooked with this method.
The researchers’ work also highlights how little we know about transgender patients with leukemia and other cancers.
Alison Alpert, MD, of the University of Rochester (N.Y.) Medical Center, and her colleagues conducted this research and presented their findings in a poster at the Acute Leukemia Forum of Hemedicus.
“There’s almost no data about transgender people with cancer ... in terms of prevalence or anything else,” Dr. Alpert noted. “And because we don’t know which patients with cancer are transgender, we can’t begin to answer any of the other big questions for patients.”
Specifically, it’s unclear what kinds of cancer transgender patients have, if there are health disparities among transgender patients, if it is safe to continue hormone therapy during cancer treatment, and if it is possible to do transition-related surgeries in the context of cancer care.
With this in mind, Dr. Alpert and her colleagues set out to identify transgender patients by detecting gender-karyotype mismatches. The team analyzed data on patients with acute myeloid leukemia (AML) or myelodysplastic syndromes enrolled in five Southwest Oncology Group (SWOG) trials.
Of the 1,748 patients analyzed, six (0.3%) had a gender-karyotype mismatch. Five patients had a 46,XY karyotype and identified as female, and one patient had a 46,XX karyotype and identified as male.
“Some transgender patients have their gender identity accurately reflected in the electronic medical record, [but] some transgender patients probably don’t,” Dr. Alpert noted. “So we identified some, but probably not all, and probably not even most, transgender patients with leukemia in this cohort.”
All six of the transgender patients identified had AML, and all were white. They ranged in age from 18 to 57 years. Four patients had achieved a complete response to therapy, and two had refractory disease.
Four patients, including one who was refractory, were still alive at last follow-up. The remaining two patients, including one who had achieved a complete response, had died.
The transgender patients identified in this analysis represent a very small percentage of the population studied, Dr. Alpert noted. Therefore, the researchers could not draw any conclusions about transgender patients with AML.
“Mostly, what we did was, we pointed out how little information we have,” Dr. Alpert said. “Oncologists don’t routinely collect gender identity information, and this information doesn’t exist in cooperative group databases either.”
“But going forward, what probably really needs to happen is that oncologists need to ask their patients whether they are transgender or not. And then, ideally, consent forms for large cooperative groups like SWOG would include gender identity data, and then we would be able to answer some of our other questions and better counsel our patients.”
Dr. Alpert and her colleagues are hoping to gain insights regarding transgender patients with lymphoma as well. The researchers are analyzing the lymphoma database at the University of Rochester Medical Center, which includes about 2,200 patients.
The team is attempting to identify transgender lymphoma patients using gender-karyotype mismatch as well as other methods, including assessing patients’ medication and surgical histories, determining whether patients have any aliases, and looking for the word “transgender” in patient charts.
“Given that the country is finally starting to talk about transgender patients, their health disparities, and their needs and experiences, it’s really time that we start collecting this data,” Dr. Alpert said.
“[I]f we are able to start to collect this data, it can help us build relationships with our patients, improve their care and outcomes, and, hopefully, be able to better counsel them about hormones and surgery.”
Dr. Alpert and her colleagues did not disclose any conflicts of interest.
The Acute Leukemia Forum is organized by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – Researchers have shown they can identify transgender leukemia patients by detecting gender-karyotype mismatches, but some transgender patients may be overlooked with this method.
The researchers’ work also highlights how little we know about transgender patients with leukemia and other cancers.
Alison Alpert, MD, of the University of Rochester (N.Y.) Medical Center, and her colleagues conducted this research and presented their findings in a poster at the Acute Leukemia Forum of Hemedicus.
“There’s almost no data about transgender people with cancer ... in terms of prevalence or anything else,” Dr. Alpert noted. “And because we don’t know which patients with cancer are transgender, we can’t begin to answer any of the other big questions for patients.”
Specifically, it’s unclear what kinds of cancer transgender patients have, if there are health disparities among transgender patients, if it is safe to continue hormone therapy during cancer treatment, and if it is possible to do transition-related surgeries in the context of cancer care.
With this in mind, Dr. Alpert and her colleagues set out to identify transgender patients by detecting gender-karyotype mismatches. The team analyzed data on patients with acute myeloid leukemia (AML) or myelodysplastic syndromes enrolled in five Southwest Oncology Group (SWOG) trials.
Of the 1,748 patients analyzed, six (0.3%) had a gender-karyotype mismatch. Five patients had a 46,XY karyotype and identified as female, and one patient had a 46,XX karyotype and identified as male.
“Some transgender patients have their gender identity accurately reflected in the electronic medical record, [but] some transgender patients probably don’t,” Dr. Alpert noted. “So we identified some, but probably not all, and probably not even most, transgender patients with leukemia in this cohort.”
All six of the transgender patients identified had AML, and all were white. They ranged in age from 18 to 57 years. Four patients had achieved a complete response to therapy, and two had refractory disease.
Four patients, including one who was refractory, were still alive at last follow-up. The remaining two patients, including one who had achieved a complete response, had died.
The transgender patients identified in this analysis represent a very small percentage of the population studied, Dr. Alpert noted. Therefore, the researchers could not draw any conclusions about transgender patients with AML.
“Mostly, what we did was, we pointed out how little information we have,” Dr. Alpert said. “Oncologists don’t routinely collect gender identity information, and this information doesn’t exist in cooperative group databases either.”
“But going forward, what probably really needs to happen is that oncologists need to ask their patients whether they are transgender or not. And then, ideally, consent forms for large cooperative groups like SWOG would include gender identity data, and then we would be able to answer some of our other questions and better counsel our patients.”
Dr. Alpert and her colleagues are hoping to gain insights regarding transgender patients with lymphoma as well. The researchers are analyzing the lymphoma database at the University of Rochester Medical Center, which includes about 2,200 patients.
The team is attempting to identify transgender lymphoma patients using gender-karyotype mismatch as well as other methods, including assessing patients’ medication and surgical histories, determining whether patients have any aliases, and looking for the word “transgender” in patient charts.
“Given that the country is finally starting to talk about transgender patients, their health disparities, and their needs and experiences, it’s really time that we start collecting this data,” Dr. Alpert said.
“[I]f we are able to start to collect this data, it can help us build relationships with our patients, improve their care and outcomes, and, hopefully, be able to better counsel them about hormones and surgery.”
Dr. Alpert and her colleagues did not disclose any conflicts of interest.
The Acute Leukemia Forum is organized by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
ALF 2019 showcases evolving treatment of AML
NEWPORT BEACH, CALIF. – The evolving treatment of acute myeloid leukemia (AML) was highlighted at the Acute Leukemia Forum of Hemedicus.
In a video interview, Martin Tallman, MD, of Memorial Sloan Kettering Cancer Center in New York, discussed several meeting presentations on the treatment of AML.
In his presentation, Craig Jordan, PhD, of the University of Colorado at Denver, Aurora, explained how the combination of venetoclax and azacitidine appears to target leukemic stem cells in AML.
Courtney DiNardo, MD, of the University of Texas MD Anderson Cancer Center, Houston, presented information on novel agents for AML, including antibody-drug conjugates; bispecific therapies; checkpoint inhibitors; and inhibitors of IDH1/2, MCL1, and MDM2.
Richard Larson, MD, of the University of Chicago, explored the possibility of an individualized approach to postremission therapy in AML.
Frederick Appelbaum, MD, of Fred Hutchinson Cancer Research Center in Seattle, showed that various maintenance therapies given after allogeneic hematopoietic stem cell transplant (HSCT) have not proven beneficial for AML patients.
Richard Jones, MD, of Johns Hopkins Medicine in Baltimore, presented data showing that post-HSCT cyclophosphamide has made haploidentical transplants safer and more effective for AML patients.
And James Ferrara, MD, of the Icahn School of Medicine at Mount Sinai, New York, detailed research showing that biomarkers of graft-versus-host disease can predict nonrelapse mortality after HSCT.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The evolving treatment of acute myeloid leukemia (AML) was highlighted at the Acute Leukemia Forum of Hemedicus.
In a video interview, Martin Tallman, MD, of Memorial Sloan Kettering Cancer Center in New York, discussed several meeting presentations on the treatment of AML.
In his presentation, Craig Jordan, PhD, of the University of Colorado at Denver, Aurora, explained how the combination of venetoclax and azacitidine appears to target leukemic stem cells in AML.
Courtney DiNardo, MD, of the University of Texas MD Anderson Cancer Center, Houston, presented information on novel agents for AML, including antibody-drug conjugates; bispecific therapies; checkpoint inhibitors; and inhibitors of IDH1/2, MCL1, and MDM2.
Richard Larson, MD, of the University of Chicago, explored the possibility of an individualized approach to postremission therapy in AML.
Frederick Appelbaum, MD, of Fred Hutchinson Cancer Research Center in Seattle, showed that various maintenance therapies given after allogeneic hematopoietic stem cell transplant (HSCT) have not proven beneficial for AML patients.
Richard Jones, MD, of Johns Hopkins Medicine in Baltimore, presented data showing that post-HSCT cyclophosphamide has made haploidentical transplants safer and more effective for AML patients.
And James Ferrara, MD, of the Icahn School of Medicine at Mount Sinai, New York, detailed research showing that biomarkers of graft-versus-host disease can predict nonrelapse mortality after HSCT.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The evolving treatment of acute myeloid leukemia (AML) was highlighted at the Acute Leukemia Forum of Hemedicus.
In a video interview, Martin Tallman, MD, of Memorial Sloan Kettering Cancer Center in New York, discussed several meeting presentations on the treatment of AML.
In his presentation, Craig Jordan, PhD, of the University of Colorado at Denver, Aurora, explained how the combination of venetoclax and azacitidine appears to target leukemic stem cells in AML.
Courtney DiNardo, MD, of the University of Texas MD Anderson Cancer Center, Houston, presented information on novel agents for AML, including antibody-drug conjugates; bispecific therapies; checkpoint inhibitors; and inhibitors of IDH1/2, MCL1, and MDM2.
Richard Larson, MD, of the University of Chicago, explored the possibility of an individualized approach to postremission therapy in AML.
Frederick Appelbaum, MD, of Fred Hutchinson Cancer Research Center in Seattle, showed that various maintenance therapies given after allogeneic hematopoietic stem cell transplant (HSCT) have not proven beneficial for AML patients.
Richard Jones, MD, of Johns Hopkins Medicine in Baltimore, presented data showing that post-HSCT cyclophosphamide has made haploidentical transplants safer and more effective for AML patients.
And James Ferrara, MD, of the Icahn School of Medicine at Mount Sinai, New York, detailed research showing that biomarkers of graft-versus-host disease can predict nonrelapse mortality after HSCT.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
Back to the drawing board for MPN combo
NEWPORT BEACH, CALIF. – The combination of ruxolitinib and decitabine will not proceed to a phase 3 trial in patients with accelerated or blast phase myeloproliferative neoplasms (MPNs).
The combination demonstrated activity and tolerability in a phase 2 trial, but outcomes were not optimal, according to Raajit K. Rampal, MD, PhD, of Memorial Sloan Kettering Cancer Center in New York.
“[P]erhaps the outcomes might be favorable compared to standard induction chemotherapy regimens,” Dr. Rampal said. “Nonetheless, it’s clear that we still have a lot of work to do, and the outcomes are not optimal in these patients.”
However, Dr. Rampal and his colleagues are investigating the possibility of combining ruxolitinib and decitabine with other agents to treat patients with accelerated or blast phase MPNs.
Dr. Rampal and his colleagues presented results from the phase 2 trial in a poster at the Acute Leukemia Forum of Hemedicus.
The trial (NCT02076191) enrolled 25 patients, 10 with accelerated phase MPN (10%-19% blasts) and 15 with blast phase MPN (at least 20% blasts). The patients’ median age was 71 years.
Patients had a median disease duration of 72.9 months. Six patients (25%) had received prior ruxolitinib, and two (8.3%) had received prior decitabine.
Treatment and safety
For the first cycle, patients received decitabine at 20 mg/m2 per day on days 8-12 and ruxolitinib at 25 mg twice a day on days 1-35. For subsequent cycles, patients received the same dose of decitabine on days 1-5 and ruxolitinib at 10 mg twice a day on days 6-28. Patients were treated until progression, withdrawal, or unacceptable toxicity.
“The adverse events we saw in this study were typical for this population, including fevers, mostly neutropenic fevers, as well as anemia and thrombocytopenia,” Dr. Rampal said.
Nonhematologic adverse events (AEs) included fatigue, abdominal pain, pneumonia, diarrhea, dizziness, and constipation. Hematologic AEs included anemia, neutropenia, febrile neutropenia, and thrombocytopenia.
Response and survival
Eighteen patients were evaluable for response. Four patients were not evaluable because they withdrew from the study due to secondary AEs and completed one cycle of therapy or less, two patients did not have circulating blasts at baseline, and one patient refused further treatment.
Among the evaluable patients, nine (50%) achieved a partial response, including four patients with accelerated phase MPN and five with blast phase MPN.
Two patients (11.1%), one with accelerated phase MPN and one with blast phase MPN, achieved a complete response with incomplete count recovery.
The remaining seven patients (38.9%), five with blast phase MPN and two with accelerated phase MPN, did not respond.
The median overall survival was 7.6 months for the entire cohort, 9.7 months for patients with blast phase MPN, and 5.8 months for patients with accelerated phase MPN.
Based on these results, Dr. Rampal and his colleagues theorized that ruxolitinib plus decitabine might be improved by the addition of other agents. The researchers are currently investigating this possibility.
“The work for this trial really came out of preclinical work in the laboratory where we combined these drugs and saw efficacy in murine models,” Dr. Rampal said. “So we’re going back to the drawing board and looking at those again to see, ‘Can we come up with new rational combinations?’ ”
Dr. Rampal and his colleagues reported having no conflicts of interest. Their study was supported by the National Institutes of Health, the National Cancer Institute, and Incyte Corporation.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The combination of ruxolitinib and decitabine will not proceed to a phase 3 trial in patients with accelerated or blast phase myeloproliferative neoplasms (MPNs).
The combination demonstrated activity and tolerability in a phase 2 trial, but outcomes were not optimal, according to Raajit K. Rampal, MD, PhD, of Memorial Sloan Kettering Cancer Center in New York.
“[P]erhaps the outcomes might be favorable compared to standard induction chemotherapy regimens,” Dr. Rampal said. “Nonetheless, it’s clear that we still have a lot of work to do, and the outcomes are not optimal in these patients.”
However, Dr. Rampal and his colleagues are investigating the possibility of combining ruxolitinib and decitabine with other agents to treat patients with accelerated or blast phase MPNs.
Dr. Rampal and his colleagues presented results from the phase 2 trial in a poster at the Acute Leukemia Forum of Hemedicus.
The trial (NCT02076191) enrolled 25 patients, 10 with accelerated phase MPN (10%-19% blasts) and 15 with blast phase MPN (at least 20% blasts). The patients’ median age was 71 years.
Patients had a median disease duration of 72.9 months. Six patients (25%) had received prior ruxolitinib, and two (8.3%) had received prior decitabine.
Treatment and safety
For the first cycle, patients received decitabine at 20 mg/m2 per day on days 8-12 and ruxolitinib at 25 mg twice a day on days 1-35. For subsequent cycles, patients received the same dose of decitabine on days 1-5 and ruxolitinib at 10 mg twice a day on days 6-28. Patients were treated until progression, withdrawal, or unacceptable toxicity.
“The adverse events we saw in this study were typical for this population, including fevers, mostly neutropenic fevers, as well as anemia and thrombocytopenia,” Dr. Rampal said.
Nonhematologic adverse events (AEs) included fatigue, abdominal pain, pneumonia, diarrhea, dizziness, and constipation. Hematologic AEs included anemia, neutropenia, febrile neutropenia, and thrombocytopenia.
Response and survival
Eighteen patients were evaluable for response. Four patients were not evaluable because they withdrew from the study due to secondary AEs and completed one cycle of therapy or less, two patients did not have circulating blasts at baseline, and one patient refused further treatment.
Among the evaluable patients, nine (50%) achieved a partial response, including four patients with accelerated phase MPN and five with blast phase MPN.
Two patients (11.1%), one with accelerated phase MPN and one with blast phase MPN, achieved a complete response with incomplete count recovery.
The remaining seven patients (38.9%), five with blast phase MPN and two with accelerated phase MPN, did not respond.
The median overall survival was 7.6 months for the entire cohort, 9.7 months for patients with blast phase MPN, and 5.8 months for patients with accelerated phase MPN.
Based on these results, Dr. Rampal and his colleagues theorized that ruxolitinib plus decitabine might be improved by the addition of other agents. The researchers are currently investigating this possibility.
“The work for this trial really came out of preclinical work in the laboratory where we combined these drugs and saw efficacy in murine models,” Dr. Rampal said. “So we’re going back to the drawing board and looking at those again to see, ‘Can we come up with new rational combinations?’ ”
Dr. Rampal and his colleagues reported having no conflicts of interest. Their study was supported by the National Institutes of Health, the National Cancer Institute, and Incyte Corporation.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
NEWPORT BEACH, CALIF. – The combination of ruxolitinib and decitabine will not proceed to a phase 3 trial in patients with accelerated or blast phase myeloproliferative neoplasms (MPNs).
The combination demonstrated activity and tolerability in a phase 2 trial, but outcomes were not optimal, according to Raajit K. Rampal, MD, PhD, of Memorial Sloan Kettering Cancer Center in New York.
“[P]erhaps the outcomes might be favorable compared to standard induction chemotherapy regimens,” Dr. Rampal said. “Nonetheless, it’s clear that we still have a lot of work to do, and the outcomes are not optimal in these patients.”
However, Dr. Rampal and his colleagues are investigating the possibility of combining ruxolitinib and decitabine with other agents to treat patients with accelerated or blast phase MPNs.
Dr. Rampal and his colleagues presented results from the phase 2 trial in a poster at the Acute Leukemia Forum of Hemedicus.
The trial (NCT02076191) enrolled 25 patients, 10 with accelerated phase MPN (10%-19% blasts) and 15 with blast phase MPN (at least 20% blasts). The patients’ median age was 71 years.
Patients had a median disease duration of 72.9 months. Six patients (25%) had received prior ruxolitinib, and two (8.3%) had received prior decitabine.
Treatment and safety
For the first cycle, patients received decitabine at 20 mg/m2 per day on days 8-12 and ruxolitinib at 25 mg twice a day on days 1-35. For subsequent cycles, patients received the same dose of decitabine on days 1-5 and ruxolitinib at 10 mg twice a day on days 6-28. Patients were treated until progression, withdrawal, or unacceptable toxicity.
“The adverse events we saw in this study were typical for this population, including fevers, mostly neutropenic fevers, as well as anemia and thrombocytopenia,” Dr. Rampal said.
Nonhematologic adverse events (AEs) included fatigue, abdominal pain, pneumonia, diarrhea, dizziness, and constipation. Hematologic AEs included anemia, neutropenia, febrile neutropenia, and thrombocytopenia.
Response and survival
Eighteen patients were evaluable for response. Four patients were not evaluable because they withdrew from the study due to secondary AEs and completed one cycle of therapy or less, two patients did not have circulating blasts at baseline, and one patient refused further treatment.
Among the evaluable patients, nine (50%) achieved a partial response, including four patients with accelerated phase MPN and five with blast phase MPN.
Two patients (11.1%), one with accelerated phase MPN and one with blast phase MPN, achieved a complete response with incomplete count recovery.
The remaining seven patients (38.9%), five with blast phase MPN and two with accelerated phase MPN, did not respond.
The median overall survival was 7.6 months for the entire cohort, 9.7 months for patients with blast phase MPN, and 5.8 months for patients with accelerated phase MPN.
Based on these results, Dr. Rampal and his colleagues theorized that ruxolitinib plus decitabine might be improved by the addition of other agents. The researchers are currently investigating this possibility.
“The work for this trial really came out of preclinical work in the laboratory where we combined these drugs and saw efficacy in murine models,” Dr. Rampal said. “So we’re going back to the drawing board and looking at those again to see, ‘Can we come up with new rational combinations?’ ”
Dr. Rampal and his colleagues reported having no conflicts of interest. Their study was supported by the National Institutes of Health, the National Cancer Institute, and Incyte Corporation.
The Acute Leukemia Forum is held by Hemedicus, which is owned by the same company as this news organization.
REPORTING FROM ALF 2019
Gilteritinib prolonged survival in FLT3-mutated AML
ATLANTA – The FLT3 inhibitor gilteritinib (Xospata) significantly prolonged overall survival, compared with salvage chemotherapy, in patients with FLT3-mutated relapsed/refractory acute myeloid leukemia, Alexander E. Perl, MD, from the Abramson Cancer Center at the University of Pennsylvania in Philadelphia reported at the 2019 annual meeting of the American Association for Cancer Research (AACR).
In a video interview, Dr. Perl discussed the results of the ADMIRAL global phase 3 randomized trial and described the current state of therapy for patients with relapsed/refractory AML bearing FLT3 mutations. Up to 70% of patients with acute myeloid leukemia will experience a relapse, and up to 40% may have disease that is resistant to induction chemotherapy. Survival for these patients is generally poor.
In particular, patients with acute myeloid leukemia and FLT3-activating mutations are at increased risk for early relapse and poor overall survival.
The ADMIRAL trial is funded by Astellas Pharma. Dr. Perl disclosed advisory board participation, consulting fees, and institutional support from Astellas and others.
ATLANTA – The FLT3 inhibitor gilteritinib (Xospata) significantly prolonged overall survival, compared with salvage chemotherapy, in patients with FLT3-mutated relapsed/refractory acute myeloid leukemia, Alexander E. Perl, MD, from the Abramson Cancer Center at the University of Pennsylvania in Philadelphia reported at the 2019 annual meeting of the American Association for Cancer Research (AACR).
In a video interview, Dr. Perl discussed the results of the ADMIRAL global phase 3 randomized trial and described the current state of therapy for patients with relapsed/refractory AML bearing FLT3 mutations. Up to 70% of patients with acute myeloid leukemia will experience a relapse, and up to 40% may have disease that is resistant to induction chemotherapy. Survival for these patients is generally poor.
In particular, patients with acute myeloid leukemia and FLT3-activating mutations are at increased risk for early relapse and poor overall survival.
The ADMIRAL trial is funded by Astellas Pharma. Dr. Perl disclosed advisory board participation, consulting fees, and institutional support from Astellas and others.
ATLANTA – The FLT3 inhibitor gilteritinib (Xospata) significantly prolonged overall survival, compared with salvage chemotherapy, in patients with FLT3-mutated relapsed/refractory acute myeloid leukemia, Alexander E. Perl, MD, from the Abramson Cancer Center at the University of Pennsylvania in Philadelphia reported at the 2019 annual meeting of the American Association for Cancer Research (AACR).
In a video interview, Dr. Perl discussed the results of the ADMIRAL global phase 3 randomized trial and described the current state of therapy for patients with relapsed/refractory AML bearing FLT3 mutations. Up to 70% of patients with acute myeloid leukemia will experience a relapse, and up to 40% may have disease that is resistant to induction chemotherapy. Survival for these patients is generally poor.
In particular, patients with acute myeloid leukemia and FLT3-activating mutations are at increased risk for early relapse and poor overall survival.
The ADMIRAL trial is funded by Astellas Pharma. Dr. Perl disclosed advisory board participation, consulting fees, and institutional support from Astellas and others.
REPORTING FROM AACR 2019
Whole-genome sequencing demonstrates clinical relevance
GLASGOW – Whole genome sequencing (WGS) appears capable of replacing cytogenetic testing and next generation sequencing (NGS) for the detection of clinically relevant molecular abnormalities in hematological malignancies, according to investigators.
A comparison of WGS with fluorescence in situ hybridization (FISH) showed that WGS caught all the same significant structural variants, plus some abnormalities that FISH had not detected, reported lead author Shirley Henderson, PhD, lead for cancer molecular diagnostics at Genomics England in Oxford.
Although further validation is needed, these findings, reported at the annual meeting of the British Society for Haematology, support an ongoing effort to validate the clinical reliability of WGS, which is currently reserved for research purposes.
“It’s vitally important that the clinical community engage with this and understand both the power and the limitations of this technique and how this work is going to be interpreted for the benefit of patients,” said Adele Fielding, PhD, session chair from University College London’s Cancer Institute.
The investigators compared WGS with FISH for detection of clinically significant structural variants (SVs) and copy number variants (CNVs) in tumor samples from 34 patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL).
The 252 standard of care FISH tests – conducted at three separate clinical diagnostic centers in the United Kingdom – included 138 SVs and 114 CNVs. WGS relied on a combination of bioinformatics and visual inspection of Circos plots. WGS confirmed all of the SVs detected by FISH with high confidence; WGS detected four additional SVs, also with high confidence, including an ETV6-RUNX1 fusion not detected by FISH because of probe limitations.
Results for CNVs were similar, with WGS detecting 78 out of 85 positive CNVs. Six of the missed positives were associated with low quality samples or low level mutations in the FISH test, suggesting that at least some positives may have been detected with better samples. Only one negative CNV from FISH was missed by WGS.
Overall, WGS had a false positive rate of less than 5% and a positive percentage agreement with FISH that exceeded 90%.
“Further work is required to fully validate all aspects of the WGS analysis pipeline,” Dr. Henderson said. “But these results indicate that WGS has the potential to reliably detect SVs and CNVs in these conditions while offering the advantage of detecting all SVs and CNVs present without the need for additional interrogation of the sample by multiple tests or probes.”
Dr. Henderson noted that there is really no “perfect method” for identifying structural and copy number variants at the present time.
Small variants are relatively easy to detect with techniques such as karyotyping and gene banding, but these tests have shortcomings, namely, that they require live cells and have “fairly high failure rates for various reasons,” Dr. Henderson said.
“FISH is an incredibly useful test and it has higher resolution than gene banding, but the problem with FISH is that you only find what you’re looking at,” Dr. Henderson said. “It’s not genome wide; it’s very targeted.”
Similarly, polymerase chain reaction (PCR), including next generation sequencing (NGS), can detect molecular abnormalities, but only those that are targeted, which may necessitate multiple tests, she said.
“If you start looking for all of the structural variants [with existing techniques], then you’re going to be doing an awful lot of tests,” Dr. Henderson said.
Another potential benefit of WGS is that it is “future resistant,” Dr. Henderson said. “As new biomarkers are discovered, you don’t have to redesign a new targeted test. It will also detect emerging biomarkers, such as mutational signatures and burden.”
The study was sponsored by NHS England. The investigators reported having no conflicts of interest.
SOURCE: Henderson S et al. BSH 2019, Abstract OR-002.
GLASGOW – Whole genome sequencing (WGS) appears capable of replacing cytogenetic testing and next generation sequencing (NGS) for the detection of clinically relevant molecular abnormalities in hematological malignancies, according to investigators.
A comparison of WGS with fluorescence in situ hybridization (FISH) showed that WGS caught all the same significant structural variants, plus some abnormalities that FISH had not detected, reported lead author Shirley Henderson, PhD, lead for cancer molecular diagnostics at Genomics England in Oxford.
Although further validation is needed, these findings, reported at the annual meeting of the British Society for Haematology, support an ongoing effort to validate the clinical reliability of WGS, which is currently reserved for research purposes.
“It’s vitally important that the clinical community engage with this and understand both the power and the limitations of this technique and how this work is going to be interpreted for the benefit of patients,” said Adele Fielding, PhD, session chair from University College London’s Cancer Institute.
The investigators compared WGS with FISH for detection of clinically significant structural variants (SVs) and copy number variants (CNVs) in tumor samples from 34 patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL).
The 252 standard of care FISH tests – conducted at three separate clinical diagnostic centers in the United Kingdom – included 138 SVs and 114 CNVs. WGS relied on a combination of bioinformatics and visual inspection of Circos plots. WGS confirmed all of the SVs detected by FISH with high confidence; WGS detected four additional SVs, also with high confidence, including an ETV6-RUNX1 fusion not detected by FISH because of probe limitations.
Results for CNVs were similar, with WGS detecting 78 out of 85 positive CNVs. Six of the missed positives were associated with low quality samples or low level mutations in the FISH test, suggesting that at least some positives may have been detected with better samples. Only one negative CNV from FISH was missed by WGS.
Overall, WGS had a false positive rate of less than 5% and a positive percentage agreement with FISH that exceeded 90%.
“Further work is required to fully validate all aspects of the WGS analysis pipeline,” Dr. Henderson said. “But these results indicate that WGS has the potential to reliably detect SVs and CNVs in these conditions while offering the advantage of detecting all SVs and CNVs present without the need for additional interrogation of the sample by multiple tests or probes.”
Dr. Henderson noted that there is really no “perfect method” for identifying structural and copy number variants at the present time.
Small variants are relatively easy to detect with techniques such as karyotyping and gene banding, but these tests have shortcomings, namely, that they require live cells and have “fairly high failure rates for various reasons,” Dr. Henderson said.
“FISH is an incredibly useful test and it has higher resolution than gene banding, but the problem with FISH is that you only find what you’re looking at,” Dr. Henderson said. “It’s not genome wide; it’s very targeted.”
Similarly, polymerase chain reaction (PCR), including next generation sequencing (NGS), can detect molecular abnormalities, but only those that are targeted, which may necessitate multiple tests, she said.
“If you start looking for all of the structural variants [with existing techniques], then you’re going to be doing an awful lot of tests,” Dr. Henderson said.
Another potential benefit of WGS is that it is “future resistant,” Dr. Henderson said. “As new biomarkers are discovered, you don’t have to redesign a new targeted test. It will also detect emerging biomarkers, such as mutational signatures and burden.”
The study was sponsored by NHS England. The investigators reported having no conflicts of interest.
SOURCE: Henderson S et al. BSH 2019, Abstract OR-002.
GLASGOW – Whole genome sequencing (WGS) appears capable of replacing cytogenetic testing and next generation sequencing (NGS) for the detection of clinically relevant molecular abnormalities in hematological malignancies, according to investigators.
A comparison of WGS with fluorescence in situ hybridization (FISH) showed that WGS caught all the same significant structural variants, plus some abnormalities that FISH had not detected, reported lead author Shirley Henderson, PhD, lead for cancer molecular diagnostics at Genomics England in Oxford.
Although further validation is needed, these findings, reported at the annual meeting of the British Society for Haematology, support an ongoing effort to validate the clinical reliability of WGS, which is currently reserved for research purposes.
“It’s vitally important that the clinical community engage with this and understand both the power and the limitations of this technique and how this work is going to be interpreted for the benefit of patients,” said Adele Fielding, PhD, session chair from University College London’s Cancer Institute.
The investigators compared WGS with FISH for detection of clinically significant structural variants (SVs) and copy number variants (CNVs) in tumor samples from 34 patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL).
The 252 standard of care FISH tests – conducted at three separate clinical diagnostic centers in the United Kingdom – included 138 SVs and 114 CNVs. WGS relied on a combination of bioinformatics and visual inspection of Circos plots. WGS confirmed all of the SVs detected by FISH with high confidence; WGS detected four additional SVs, also with high confidence, including an ETV6-RUNX1 fusion not detected by FISH because of probe limitations.
Results for CNVs were similar, with WGS detecting 78 out of 85 positive CNVs. Six of the missed positives were associated with low quality samples or low level mutations in the FISH test, suggesting that at least some positives may have been detected with better samples. Only one negative CNV from FISH was missed by WGS.
Overall, WGS had a false positive rate of less than 5% and a positive percentage agreement with FISH that exceeded 90%.
“Further work is required to fully validate all aspects of the WGS analysis pipeline,” Dr. Henderson said. “But these results indicate that WGS has the potential to reliably detect SVs and CNVs in these conditions while offering the advantage of detecting all SVs and CNVs present without the need for additional interrogation of the sample by multiple tests or probes.”
Dr. Henderson noted that there is really no “perfect method” for identifying structural and copy number variants at the present time.
Small variants are relatively easy to detect with techniques such as karyotyping and gene banding, but these tests have shortcomings, namely, that they require live cells and have “fairly high failure rates for various reasons,” Dr. Henderson said.
“FISH is an incredibly useful test and it has higher resolution than gene banding, but the problem with FISH is that you only find what you’re looking at,” Dr. Henderson said. “It’s not genome wide; it’s very targeted.”
Similarly, polymerase chain reaction (PCR), including next generation sequencing (NGS), can detect molecular abnormalities, but only those that are targeted, which may necessitate multiple tests, she said.
“If you start looking for all of the structural variants [with existing techniques], then you’re going to be doing an awful lot of tests,” Dr. Henderson said.
Another potential benefit of WGS is that it is “future resistant,” Dr. Henderson said. “As new biomarkers are discovered, you don’t have to redesign a new targeted test. It will also detect emerging biomarkers, such as mutational signatures and burden.”
The study was sponsored by NHS England. The investigators reported having no conflicts of interest.
SOURCE: Henderson S et al. BSH 2019, Abstract OR-002.
REPORTING FROM BSH 2019








