User login
New nanoparticles may improve chemo delivery
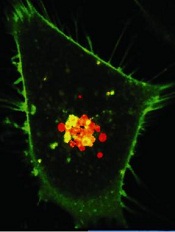
Image from PNAS
A new type of nanoparticle can deliver chemotherapy directly and efficiently to individual cells, according to research published in the Journal of the American Chemical Society.
These nanoparticles, known as connectosomes, are equipped with gap junctions—a pathway that allows for the rapid movement of molecules between 2 cells.
The gap junctions allow the connectosomes to create a direct channel to deliver drugs to each individual cell.
“Gap junctions are the cells’ mechanism for sharing small molecules between neighboring cells,” said study author Jeanne Stachowiak, PhD, of The University of Texas at Austin.
“We believed that there must be a way to utilize them for better drug delivery. The big challenge was in making the materials efficiently and showing that the drugs are delivered through the gap junctions and not some other component.”
To create the connectosomes, the researchers used a chemical process to derive liposomes from donor cells that were engineered to over-produce gap junctions, which are made of proteins.
The team then loaded the connectosomes with the chemotherapy drug doxorubicin.
In in vitro tests with human cells, the researchers found that doxorubicin delivered through connectosomes was 10 times as efficient at killing cancer cells as freely delivered doxorubicin.
Connectosomes were also 100 to 100,000 times as efficient as conventional nanoparticles in delivering doxorubicin, because a drug can diffuse more efficiently through a gap junction than across the oily lipid membrane.
“Connectosomes could open doors for the improved utilization of nanoparticles to deliver other types of therapies,” said Avinash Gadok, a doctoral student at The University of Texas at Austin.
“A huge advantage of nanoparticles is that they can target cells, which helps protect off-target tissues.”
Now, the researchers are investigating whether connectosomes can biochemically target tumor cells and whether they could be useful in inhibiting the migration of tumor cells.
Gap junctions are known to suppress cell migration, creating the potential for connectosomes to help control the movement of tumor cells out of the tumor and into the bloodstream.
“We would like to see whether this approach could delay metastasis while treating the tumor,” Dr Stachowiak said.
“It would be nice to have a multi-pronged approach where you have a particle that slows down metastasis, rapidly delivers drugs, and turns off expression of genes that are promoting the migration of tumor cells.” ![]()

Image from PNAS
A new type of nanoparticle can deliver chemotherapy directly and efficiently to individual cells, according to research published in the Journal of the American Chemical Society.
These nanoparticles, known as connectosomes, are equipped with gap junctions—a pathway that allows for the rapid movement of molecules between 2 cells.
The gap junctions allow the connectosomes to create a direct channel to deliver drugs to each individual cell.
“Gap junctions are the cells’ mechanism for sharing small molecules between neighboring cells,” said study author Jeanne Stachowiak, PhD, of The University of Texas at Austin.
“We believed that there must be a way to utilize them for better drug delivery. The big challenge was in making the materials efficiently and showing that the drugs are delivered through the gap junctions and not some other component.”
To create the connectosomes, the researchers used a chemical process to derive liposomes from donor cells that were engineered to over-produce gap junctions, which are made of proteins.
The team then loaded the connectosomes with the chemotherapy drug doxorubicin.
In in vitro tests with human cells, the researchers found that doxorubicin delivered through connectosomes was 10 times as efficient at killing cancer cells as freely delivered doxorubicin.
Connectosomes were also 100 to 100,000 times as efficient as conventional nanoparticles in delivering doxorubicin, because a drug can diffuse more efficiently through a gap junction than across the oily lipid membrane.
“Connectosomes could open doors for the improved utilization of nanoparticles to deliver other types of therapies,” said Avinash Gadok, a doctoral student at The University of Texas at Austin.
“A huge advantage of nanoparticles is that they can target cells, which helps protect off-target tissues.”
Now, the researchers are investigating whether connectosomes can biochemically target tumor cells and whether they could be useful in inhibiting the migration of tumor cells.
Gap junctions are known to suppress cell migration, creating the potential for connectosomes to help control the movement of tumor cells out of the tumor and into the bloodstream.
“We would like to see whether this approach could delay metastasis while treating the tumor,” Dr Stachowiak said.
“It would be nice to have a multi-pronged approach where you have a particle that slows down metastasis, rapidly delivers drugs, and turns off expression of genes that are promoting the migration of tumor cells.” ![]()

Image from PNAS
A new type of nanoparticle can deliver chemotherapy directly and efficiently to individual cells, according to research published in the Journal of the American Chemical Society.
These nanoparticles, known as connectosomes, are equipped with gap junctions—a pathway that allows for the rapid movement of molecules between 2 cells.
The gap junctions allow the connectosomes to create a direct channel to deliver drugs to each individual cell.
“Gap junctions are the cells’ mechanism for sharing small molecules between neighboring cells,” said study author Jeanne Stachowiak, PhD, of The University of Texas at Austin.
“We believed that there must be a way to utilize them for better drug delivery. The big challenge was in making the materials efficiently and showing that the drugs are delivered through the gap junctions and not some other component.”
To create the connectosomes, the researchers used a chemical process to derive liposomes from donor cells that were engineered to over-produce gap junctions, which are made of proteins.
The team then loaded the connectosomes with the chemotherapy drug doxorubicin.
In in vitro tests with human cells, the researchers found that doxorubicin delivered through connectosomes was 10 times as efficient at killing cancer cells as freely delivered doxorubicin.
Connectosomes were also 100 to 100,000 times as efficient as conventional nanoparticles in delivering doxorubicin, because a drug can diffuse more efficiently through a gap junction than across the oily lipid membrane.
“Connectosomes could open doors for the improved utilization of nanoparticles to deliver other types of therapies,” said Avinash Gadok, a doctoral student at The University of Texas at Austin.
“A huge advantage of nanoparticles is that they can target cells, which helps protect off-target tissues.”
Now, the researchers are investigating whether connectosomes can biochemically target tumor cells and whether they could be useful in inhibiting the migration of tumor cells.
Gap junctions are known to suppress cell migration, creating the potential for connectosomes to help control the movement of tumor cells out of the tumor and into the bloodstream.
“We would like to see whether this approach could delay metastasis while treating the tumor,” Dr Stachowiak said.
“It would be nice to have a multi-pronged approach where you have a particle that slows down metastasis, rapidly delivers drugs, and turns off expression of genes that are promoting the migration of tumor cells.” ![]()
How a protein regulates blood cell fate
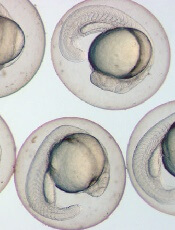
Image by Ian Johnston
Results of preclinical research explain the role endoglin plays in hematopoietic cell fate during embryogenesis.
Previous research showed that endoglin, a cell surface protein belonging to the TGF-beta receptor complex, is required for early hematopoietic lineage specification.
A new study shows that endoglin modulates the BMP and Wnt signaling pathways to encourage progenitor cells to develop into blood cells rather than cardiac cells.
This study was published in Nature Communications.
“During the early stages of development, cells have to make decisions very quickly,” said study author Rita Perlingeiro, PhD, of the University of Minnesota in Minneapolis.
“Fine-tuning of these early cell fate decisions can be easily disrupted by levels of key proteins within these cells. When one cell type is favored, this implies less of another. In this case, high levels of endoglin expression enhance the cell differentiation into blood cells, whereas cardiac cells are in deficit.”
Dr Perlingeiro and her colleagues made this discovery studying zebrafish and mouse models. The team wanted to pinpoint the mechanism underlying the dual function of endoglin in blood cell and cardiac cell fate.
The researchers found that endoglin is expressed in early mesoderm, and it marks both hematopoietic and cardiac progenitors.
Experiments showed that high levels of endoglin increase hematopoiesis while inhibiting cardiogenesis. And the levels of endoglin determine the activation of the BMP and Wnt signaling pathways.
With further investigation, the researchers identified JDP2, a member of the AP-1 transcription factor family, as an endoglin-dependent downstream target of Wnt signaling.
The team found that JDP2 expression is sufficient to establish blood cell fate when BMP and Wnt crosstalk is disturbed.
“The blood and heart systems are the first organs to develop in mammals, but the mechanisms regulating these earliest cell fate decisions are poorly understood,” Dr Perlingeiro said.
“By using multiple model systems, combined with specialized cell sorting technology and sequencing tools, our findings help uncover mechanisms previously unseen in the few cells engaged in these early development decisions.” ![]()

Image by Ian Johnston
Results of preclinical research explain the role endoglin plays in hematopoietic cell fate during embryogenesis.
Previous research showed that endoglin, a cell surface protein belonging to the TGF-beta receptor complex, is required for early hematopoietic lineage specification.
A new study shows that endoglin modulates the BMP and Wnt signaling pathways to encourage progenitor cells to develop into blood cells rather than cardiac cells.
This study was published in Nature Communications.
“During the early stages of development, cells have to make decisions very quickly,” said study author Rita Perlingeiro, PhD, of the University of Minnesota in Minneapolis.
“Fine-tuning of these early cell fate decisions can be easily disrupted by levels of key proteins within these cells. When one cell type is favored, this implies less of another. In this case, high levels of endoglin expression enhance the cell differentiation into blood cells, whereas cardiac cells are in deficit.”
Dr Perlingeiro and her colleagues made this discovery studying zebrafish and mouse models. The team wanted to pinpoint the mechanism underlying the dual function of endoglin in blood cell and cardiac cell fate.
The researchers found that endoglin is expressed in early mesoderm, and it marks both hematopoietic and cardiac progenitors.
Experiments showed that high levels of endoglin increase hematopoiesis while inhibiting cardiogenesis. And the levels of endoglin determine the activation of the BMP and Wnt signaling pathways.
With further investigation, the researchers identified JDP2, a member of the AP-1 transcription factor family, as an endoglin-dependent downstream target of Wnt signaling.
The team found that JDP2 expression is sufficient to establish blood cell fate when BMP and Wnt crosstalk is disturbed.
“The blood and heart systems are the first organs to develop in mammals, but the mechanisms regulating these earliest cell fate decisions are poorly understood,” Dr Perlingeiro said.
“By using multiple model systems, combined with specialized cell sorting technology and sequencing tools, our findings help uncover mechanisms previously unseen in the few cells engaged in these early development decisions.” ![]()

Image by Ian Johnston
Results of preclinical research explain the role endoglin plays in hematopoietic cell fate during embryogenesis.
Previous research showed that endoglin, a cell surface protein belonging to the TGF-beta receptor complex, is required for early hematopoietic lineage specification.
A new study shows that endoglin modulates the BMP and Wnt signaling pathways to encourage progenitor cells to develop into blood cells rather than cardiac cells.
This study was published in Nature Communications.
“During the early stages of development, cells have to make decisions very quickly,” said study author Rita Perlingeiro, PhD, of the University of Minnesota in Minneapolis.
“Fine-tuning of these early cell fate decisions can be easily disrupted by levels of key proteins within these cells. When one cell type is favored, this implies less of another. In this case, high levels of endoglin expression enhance the cell differentiation into blood cells, whereas cardiac cells are in deficit.”
Dr Perlingeiro and her colleagues made this discovery studying zebrafish and mouse models. The team wanted to pinpoint the mechanism underlying the dual function of endoglin in blood cell and cardiac cell fate.
The researchers found that endoglin is expressed in early mesoderm, and it marks both hematopoietic and cardiac progenitors.
Experiments showed that high levels of endoglin increase hematopoiesis while inhibiting cardiogenesis. And the levels of endoglin determine the activation of the BMP and Wnt signaling pathways.
With further investigation, the researchers identified JDP2, a member of the AP-1 transcription factor family, as an endoglin-dependent downstream target of Wnt signaling.
The team found that JDP2 expression is sufficient to establish blood cell fate when BMP and Wnt crosstalk is disturbed.
“The blood and heart systems are the first organs to develop in mammals, but the mechanisms regulating these earliest cell fate decisions are poorly understood,” Dr Perlingeiro said.
“By using multiple model systems, combined with specialized cell sorting technology and sequencing tools, our findings help uncover mechanisms previously unseen in the few cells engaged in these early development decisions.” ![]()
Holographic imaging and deep learning diagnose malaria
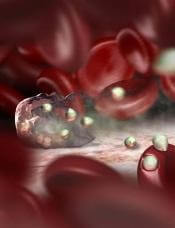
Image by Peter H. Seeberger
Scientists say they have devised a technique that can be used to diagnose malaria quickly and with clinically relevant accuracy.
The technique involves using computer deep learning and light-based, holographic scans to spot malaria-infected cells from an untouched blood sample without any help from a human.
The scientists believe this could form the basis of a fast, reliable malaria test that could be given by most anyone, anywhere in the field.
The team described the method in PLOS ONE.
“With this technique, the path is there to be able to process thousands of cells per minute,” said study author Adam Wax, PhD, of Duke University in Durham, North Carolina.
“That’s a huge improvement to the 40 minutes it currently takes a field technician to stain, prepare, and read a slide to personally look for infection.”
The new technique is based on a technology called quantitative phase spectroscopy. As a laser sweeps through the visible spectrum of light, sensors capture how each discrete light frequency interacts with a sample of blood.
The resulting data captures a holographic image that provides a wide array of information that can indicate a malaria infection.
“We identified 23 parameters that are statistically significant for spotting malaria,” said study author Han Sang Park, a doctoral student in Dr Wax’s lab.
For example, as the disease progresses, red blood cells decrease in volume, lose hemoglobin, and deform as the parasite within grows larger. This affects features such as cell volume, perimeter, shape, and center of mass.
“However, none of the parameters were reliable more than 90% of the time on their own,” Park said. “So we decided to use them all.”
“To be adopted, any new diagnostic device has to be just as reliable as a trained field worker with a microscope,” Dr Wax said. “Otherwise, even with a 90% success rate, you’d still miss more than 20 million [malaria] cases a year.”
To get a more accurate reading, Dr Wax and his colleagues turned to deep learning—a method by which computers teach themselves how to distinguish between different objects.
Feeding data on healthy and diseased cells into a computer enabled the deep learning program to determine which sets of measurements at which thresholds most clearly distinguished healthy cells from diseased cells.
When the scientists put the resulting algorithm to the test with hundreds of cells, the algorithm was able to correctly spot malaria more than 95% of the time—a number the team believes will increase as more cells are used to train the program.
The team noted that, because the technique breaks data-rich holograms down to just 23 numbers, tests can be easily transmitted in bulk. They said this is important for locations that often do not have reliable, fast internet connections.
Dr Wax and his colleagues are now looking to develop the technology into a diagnostic device through a startup company called M2 Photonics Innovations. They hope to show that a device based on this technology would be accurate and cost-efficient enough to be useful in the field.
Dr Wax has also received funding to begin exploring the use of the technique for spotting cancerous cells in blood samples. ![]()

Image by Peter H. Seeberger
Scientists say they have devised a technique that can be used to diagnose malaria quickly and with clinically relevant accuracy.
The technique involves using computer deep learning and light-based, holographic scans to spot malaria-infected cells from an untouched blood sample without any help from a human.
The scientists believe this could form the basis of a fast, reliable malaria test that could be given by most anyone, anywhere in the field.
The team described the method in PLOS ONE.
“With this technique, the path is there to be able to process thousands of cells per minute,” said study author Adam Wax, PhD, of Duke University in Durham, North Carolina.
“That’s a huge improvement to the 40 minutes it currently takes a field technician to stain, prepare, and read a slide to personally look for infection.”
The new technique is based on a technology called quantitative phase spectroscopy. As a laser sweeps through the visible spectrum of light, sensors capture how each discrete light frequency interacts with a sample of blood.
The resulting data captures a holographic image that provides a wide array of information that can indicate a malaria infection.
“We identified 23 parameters that are statistically significant for spotting malaria,” said study author Han Sang Park, a doctoral student in Dr Wax’s lab.
For example, as the disease progresses, red blood cells decrease in volume, lose hemoglobin, and deform as the parasite within grows larger. This affects features such as cell volume, perimeter, shape, and center of mass.
“However, none of the parameters were reliable more than 90% of the time on their own,” Park said. “So we decided to use them all.”
“To be adopted, any new diagnostic device has to be just as reliable as a trained field worker with a microscope,” Dr Wax said. “Otherwise, even with a 90% success rate, you’d still miss more than 20 million [malaria] cases a year.”
To get a more accurate reading, Dr Wax and his colleagues turned to deep learning—a method by which computers teach themselves how to distinguish between different objects.
Feeding data on healthy and diseased cells into a computer enabled the deep learning program to determine which sets of measurements at which thresholds most clearly distinguished healthy cells from diseased cells.
When the scientists put the resulting algorithm to the test with hundreds of cells, the algorithm was able to correctly spot malaria more than 95% of the time—a number the team believes will increase as more cells are used to train the program.
The team noted that, because the technique breaks data-rich holograms down to just 23 numbers, tests can be easily transmitted in bulk. They said this is important for locations that often do not have reliable, fast internet connections.
Dr Wax and his colleagues are now looking to develop the technology into a diagnostic device through a startup company called M2 Photonics Innovations. They hope to show that a device based on this technology would be accurate and cost-efficient enough to be useful in the field.
Dr Wax has also received funding to begin exploring the use of the technique for spotting cancerous cells in blood samples. ![]()

Image by Peter H. Seeberger
Scientists say they have devised a technique that can be used to diagnose malaria quickly and with clinically relevant accuracy.
The technique involves using computer deep learning and light-based, holographic scans to spot malaria-infected cells from an untouched blood sample without any help from a human.
The scientists believe this could form the basis of a fast, reliable malaria test that could be given by most anyone, anywhere in the field.
The team described the method in PLOS ONE.
“With this technique, the path is there to be able to process thousands of cells per minute,” said study author Adam Wax, PhD, of Duke University in Durham, North Carolina.
“That’s a huge improvement to the 40 minutes it currently takes a field technician to stain, prepare, and read a slide to personally look for infection.”
The new technique is based on a technology called quantitative phase spectroscopy. As a laser sweeps through the visible spectrum of light, sensors capture how each discrete light frequency interacts with a sample of blood.
The resulting data captures a holographic image that provides a wide array of information that can indicate a malaria infection.
“We identified 23 parameters that are statistically significant for spotting malaria,” said study author Han Sang Park, a doctoral student in Dr Wax’s lab.
For example, as the disease progresses, red blood cells decrease in volume, lose hemoglobin, and deform as the parasite within grows larger. This affects features such as cell volume, perimeter, shape, and center of mass.
“However, none of the parameters were reliable more than 90% of the time on their own,” Park said. “So we decided to use them all.”
“To be adopted, any new diagnostic device has to be just as reliable as a trained field worker with a microscope,” Dr Wax said. “Otherwise, even with a 90% success rate, you’d still miss more than 20 million [malaria] cases a year.”
To get a more accurate reading, Dr Wax and his colleagues turned to deep learning—a method by which computers teach themselves how to distinguish between different objects.
Feeding data on healthy and diseased cells into a computer enabled the deep learning program to determine which sets of measurements at which thresholds most clearly distinguished healthy cells from diseased cells.
When the scientists put the resulting algorithm to the test with hundreds of cells, the algorithm was able to correctly spot malaria more than 95% of the time—a number the team believes will increase as more cells are used to train the program.
The team noted that, because the technique breaks data-rich holograms down to just 23 numbers, tests can be easily transmitted in bulk. They said this is important for locations that often do not have reliable, fast internet connections.
Dr Wax and his colleagues are now looking to develop the technology into a diagnostic device through a startup company called M2 Photonics Innovations. They hope to show that a device based on this technology would be accurate and cost-efficient enough to be useful in the field.
Dr Wax has also received funding to begin exploring the use of the technique for spotting cancerous cells in blood samples. ![]()
Findings may aid drug delivery, bioimaging

to illuminate microfluidic device
simulating a blood vessel.
Photo from Anson Ma/UConn
A study published in Biophysical Journal has revealed new information about how particles behave in the bloodstream, and investigators believe the findings have implications for bioimaging and targeted drug delivery in cancer.
The investigators used a microfluidic channel device to observe, track, and measure how individual particles behave in a simulated blood vessel.
Their goal was to learn more about the physics influencing a particle’s behavior as it travels in the blood and to determine which particle size might be the most effective for delivering drugs to their targets.
“Even before particles reach a target site, you have to worry about what is going to happen with them after they get injected into the bloodstream,” said study author Anson Ma, PhD, of the University of Connecticut in Storrs, Connecticut.
“Are they going to clump together? How are they going to move around? Are they going to get swept away and flushed out of our bodies?”
Using a high-powered fluorescence microscope, Dr Ma and his colleagues were able to observe particles being carried along in the simulated blood vessel in what could be described as a vascular “Running of the Bulls.”
Red blood cells raced through the middle of the channel, and the particles were carried along in the rush, bumping and bouncing off the blood cells until they were pushed to open spaces—called the cell-free layer—along the vessel’s walls.
The investigators found that larger particles—the optimum size appeared to be about 2 microns—were most likely to get pushed to the cell-free layer, where their chances of carrying a drug to a targeted site are greatest.
The team also determined that 2 microns was the largest size that should be used if particles are going to have any chance of going through the leaky blood vessel walls to the site.
“When it comes to using particles for the delivery of cancer drugs, size matters,” Dr Ma said. “When you have a bigger particle, the chance of it bumping into blood cells is much higher, there are a lot more collisions, and they tend to get pushed to the blood vessel walls.”
These results were somewhat surprising. The investigators had theorized that smaller particles would probably be the most effective since they would move the most in collisions with blood cells.
But the opposite proved true. The smaller particles appeared to skirt through the mass of moving blood cells and were less likely to get bounced to the cell-free layer.
Knowing how particles behave in the circulatory system should help improve targeted drug delivery, Dr Ma said. And this should further reduce the side effects caused by potent cancer drugs missing their target.
Measuring how different sized particles move in the bloodstream may also be beneficial in bioimaging, where the goal is to keep particles circulating in the bloodstream long enough for imaging to occur. In that case, smaller particles would be better, Dr Ma said.
Moving forward, Dr Ma would like to explore other aspects of particle flow in the circulatory system, such as how particles behave when they pass through a constricted area, like from a blood vessel to a capillary.
Capillaries are only about 7 microns in diameter. Dr Ma said he would like to know how that constricted space might impact particle flow or the ability of particles to accumulate near the vessel walls.
“We have all of this complex geometry in our bodies,” Dr Ma said. “Most people just assume there is no impact when a particle moves from a bigger channel to a smaller channel because they haven’t quantified it. Our plan is to do some experiments to look at this more carefully, building on the work that we just published.” ![]()

to illuminate microfluidic device
simulating a blood vessel.
Photo from Anson Ma/UConn
A study published in Biophysical Journal has revealed new information about how particles behave in the bloodstream, and investigators believe the findings have implications for bioimaging and targeted drug delivery in cancer.
The investigators used a microfluidic channel device to observe, track, and measure how individual particles behave in a simulated blood vessel.
Their goal was to learn more about the physics influencing a particle’s behavior as it travels in the blood and to determine which particle size might be the most effective for delivering drugs to their targets.
“Even before particles reach a target site, you have to worry about what is going to happen with them after they get injected into the bloodstream,” said study author Anson Ma, PhD, of the University of Connecticut in Storrs, Connecticut.
“Are they going to clump together? How are they going to move around? Are they going to get swept away and flushed out of our bodies?”
Using a high-powered fluorescence microscope, Dr Ma and his colleagues were able to observe particles being carried along in the simulated blood vessel in what could be described as a vascular “Running of the Bulls.”
Red blood cells raced through the middle of the channel, and the particles were carried along in the rush, bumping and bouncing off the blood cells until they were pushed to open spaces—called the cell-free layer—along the vessel’s walls.
The investigators found that larger particles—the optimum size appeared to be about 2 microns—were most likely to get pushed to the cell-free layer, where their chances of carrying a drug to a targeted site are greatest.
The team also determined that 2 microns was the largest size that should be used if particles are going to have any chance of going through the leaky blood vessel walls to the site.
“When it comes to using particles for the delivery of cancer drugs, size matters,” Dr Ma said. “When you have a bigger particle, the chance of it bumping into blood cells is much higher, there are a lot more collisions, and they tend to get pushed to the blood vessel walls.”
These results were somewhat surprising. The investigators had theorized that smaller particles would probably be the most effective since they would move the most in collisions with blood cells.
But the opposite proved true. The smaller particles appeared to skirt through the mass of moving blood cells and were less likely to get bounced to the cell-free layer.
Knowing how particles behave in the circulatory system should help improve targeted drug delivery, Dr Ma said. And this should further reduce the side effects caused by potent cancer drugs missing their target.
Measuring how different sized particles move in the bloodstream may also be beneficial in bioimaging, where the goal is to keep particles circulating in the bloodstream long enough for imaging to occur. In that case, smaller particles would be better, Dr Ma said.
Moving forward, Dr Ma would like to explore other aspects of particle flow in the circulatory system, such as how particles behave when they pass through a constricted area, like from a blood vessel to a capillary.
Capillaries are only about 7 microns in diameter. Dr Ma said he would like to know how that constricted space might impact particle flow or the ability of particles to accumulate near the vessel walls.
“We have all of this complex geometry in our bodies,” Dr Ma said. “Most people just assume there is no impact when a particle moves from a bigger channel to a smaller channel because they haven’t quantified it. Our plan is to do some experiments to look at this more carefully, building on the work that we just published.” ![]()

to illuminate microfluidic device
simulating a blood vessel.
Photo from Anson Ma/UConn
A study published in Biophysical Journal has revealed new information about how particles behave in the bloodstream, and investigators believe the findings have implications for bioimaging and targeted drug delivery in cancer.
The investigators used a microfluidic channel device to observe, track, and measure how individual particles behave in a simulated blood vessel.
Their goal was to learn more about the physics influencing a particle’s behavior as it travels in the blood and to determine which particle size might be the most effective for delivering drugs to their targets.
“Even before particles reach a target site, you have to worry about what is going to happen with them after they get injected into the bloodstream,” said study author Anson Ma, PhD, of the University of Connecticut in Storrs, Connecticut.
“Are they going to clump together? How are they going to move around? Are they going to get swept away and flushed out of our bodies?”
Using a high-powered fluorescence microscope, Dr Ma and his colleagues were able to observe particles being carried along in the simulated blood vessel in what could be described as a vascular “Running of the Bulls.”
Red blood cells raced through the middle of the channel, and the particles were carried along in the rush, bumping and bouncing off the blood cells until they were pushed to open spaces—called the cell-free layer—along the vessel’s walls.
The investigators found that larger particles—the optimum size appeared to be about 2 microns—were most likely to get pushed to the cell-free layer, where their chances of carrying a drug to a targeted site are greatest.
The team also determined that 2 microns was the largest size that should be used if particles are going to have any chance of going through the leaky blood vessel walls to the site.
“When it comes to using particles for the delivery of cancer drugs, size matters,” Dr Ma said. “When you have a bigger particle, the chance of it bumping into blood cells is much higher, there are a lot more collisions, and they tend to get pushed to the blood vessel walls.”
These results were somewhat surprising. The investigators had theorized that smaller particles would probably be the most effective since they would move the most in collisions with blood cells.
But the opposite proved true. The smaller particles appeared to skirt through the mass of moving blood cells and were less likely to get bounced to the cell-free layer.
Knowing how particles behave in the circulatory system should help improve targeted drug delivery, Dr Ma said. And this should further reduce the side effects caused by potent cancer drugs missing their target.
Measuring how different sized particles move in the bloodstream may also be beneficial in bioimaging, where the goal is to keep particles circulating in the bloodstream long enough for imaging to occur. In that case, smaller particles would be better, Dr Ma said.
Moving forward, Dr Ma would like to explore other aspects of particle flow in the circulatory system, such as how particles behave when they pass through a constricted area, like from a blood vessel to a capillary.
Capillaries are only about 7 microns in diameter. Dr Ma said he would like to know how that constricted space might impact particle flow or the ability of particles to accumulate near the vessel walls.
“We have all of this complex geometry in our bodies,” Dr Ma said. “Most people just assume there is no impact when a particle moves from a bigger channel to a smaller channel because they haven’t quantified it. Our plan is to do some experiments to look at this more carefully, building on the work that we just published.” ![]()
Scientist awarded Nobel Prize for autophagy research

Photo by Mari Honda
The 2016 Nobel Prize in Physiology or Medicine has been awarded to Yoshinori Ohsumi, PhD, for his discoveries related to autophagy.
The concept of autophagy emerged during the 1960s, but little was known about the process until the early 1990s.
That’s when Dr Ohsumi used yeast cells to identify genes essential for autophagy. He cloned several of these genes in yeast and mammalian cells and described the function of the encoded proteins.
According to The Nobel Assembly at Karolinska Institutet, Dr Ohsumi’s discoveries opened the path to understanding the fundamental importance of autophagy in many physiological processes.
The man
Dr Ohsumi was born in 1945 in Fukuoka, Japan. He received a PhD from University of Tokyo in 1974.
After spending 3 years at Rockefeller University in New York, he returned to the University of Tokyo, where he established his research group in 1988. Since 2009, he has been a professor at the Tokyo Institute of Technology.
The research
The Belgian scientist Christian de Duve coined the term autophagy in 1963. However, the process was still not well understood when Dr Ohsumi began his research on autophagy.
In the early 1990s, Dr Ohsumi decided to study autophagy using the budding yeast Saccharomyces cerevisae as a model system.
He was not sure whether autophagy existed in this organism. However, he reasoned that, if it did, and he could disrupt the degradation process in the vacuole while autophagy was active, autophagosomes should accumulate within the vacuole.
Dr Ohsumi cultured mutated yeast lacking vacuolar degradation enzymes and simultaneously stimulated autophagy by starving the cells.
Within hours, the vacuoles were filled with small vesicles that had not been degraded. The vesicles were autophagosomes, and the experiment proved that autophagy exists in yeast cells.
The experiment also provided a method to identify and characterize genes involved in autophagy.
Dr Ohsumi noted that the accumulation of autophagosomes should not occur if genes important for autophagy were inactivated.
So he exposed the yeast cells to a chemical that randomly introduced mutations in many genes, and then he induced autophagy. In this way, he identified 15 genes essential for autophagy in budding yeast.
In his subsequent studies, Dr Ohsumi cloned several of these genes in yeast and mammalian cells and characterized the function of the proteins encoded by these genes.
He found that autophagy is controlled by a cascade of proteins and protein complexes, each regulating a distinct stage of autophagosome initiation and formation.
Insights provided by Dr Ohsumi’s work enabled subsequent research that has revealed the role of autophagy in human physiology and disease.
For more information on Dr Ohsumi and his work, visit the Nobel Prize website. ![]()

Photo by Mari Honda
The 2016 Nobel Prize in Physiology or Medicine has been awarded to Yoshinori Ohsumi, PhD, for his discoveries related to autophagy.
The concept of autophagy emerged during the 1960s, but little was known about the process until the early 1990s.
That’s when Dr Ohsumi used yeast cells to identify genes essential for autophagy. He cloned several of these genes in yeast and mammalian cells and described the function of the encoded proteins.
According to The Nobel Assembly at Karolinska Institutet, Dr Ohsumi’s discoveries opened the path to understanding the fundamental importance of autophagy in many physiological processes.
The man
Dr Ohsumi was born in 1945 in Fukuoka, Japan. He received a PhD from University of Tokyo in 1974.
After spending 3 years at Rockefeller University in New York, he returned to the University of Tokyo, where he established his research group in 1988. Since 2009, he has been a professor at the Tokyo Institute of Technology.
The research
The Belgian scientist Christian de Duve coined the term autophagy in 1963. However, the process was still not well understood when Dr Ohsumi began his research on autophagy.
In the early 1990s, Dr Ohsumi decided to study autophagy using the budding yeast Saccharomyces cerevisae as a model system.
He was not sure whether autophagy existed in this organism. However, he reasoned that, if it did, and he could disrupt the degradation process in the vacuole while autophagy was active, autophagosomes should accumulate within the vacuole.
Dr Ohsumi cultured mutated yeast lacking vacuolar degradation enzymes and simultaneously stimulated autophagy by starving the cells.
Within hours, the vacuoles were filled with small vesicles that had not been degraded. The vesicles were autophagosomes, and the experiment proved that autophagy exists in yeast cells.
The experiment also provided a method to identify and characterize genes involved in autophagy.
Dr Ohsumi noted that the accumulation of autophagosomes should not occur if genes important for autophagy were inactivated.
So he exposed the yeast cells to a chemical that randomly introduced mutations in many genes, and then he induced autophagy. In this way, he identified 15 genes essential for autophagy in budding yeast.
In his subsequent studies, Dr Ohsumi cloned several of these genes in yeast and mammalian cells and characterized the function of the proteins encoded by these genes.
He found that autophagy is controlled by a cascade of proteins and protein complexes, each regulating a distinct stage of autophagosome initiation and formation.
Insights provided by Dr Ohsumi’s work enabled subsequent research that has revealed the role of autophagy in human physiology and disease.
For more information on Dr Ohsumi and his work, visit the Nobel Prize website. ![]()

Photo by Mari Honda
The 2016 Nobel Prize in Physiology or Medicine has been awarded to Yoshinori Ohsumi, PhD, for his discoveries related to autophagy.
The concept of autophagy emerged during the 1960s, but little was known about the process until the early 1990s.
That’s when Dr Ohsumi used yeast cells to identify genes essential for autophagy. He cloned several of these genes in yeast and mammalian cells and described the function of the encoded proteins.
According to The Nobel Assembly at Karolinska Institutet, Dr Ohsumi’s discoveries opened the path to understanding the fundamental importance of autophagy in many physiological processes.
The man
Dr Ohsumi was born in 1945 in Fukuoka, Japan. He received a PhD from University of Tokyo in 1974.
After spending 3 years at Rockefeller University in New York, he returned to the University of Tokyo, where he established his research group in 1988. Since 2009, he has been a professor at the Tokyo Institute of Technology.
The research
The Belgian scientist Christian de Duve coined the term autophagy in 1963. However, the process was still not well understood when Dr Ohsumi began his research on autophagy.
In the early 1990s, Dr Ohsumi decided to study autophagy using the budding yeast Saccharomyces cerevisae as a model system.
He was not sure whether autophagy existed in this organism. However, he reasoned that, if it did, and he could disrupt the degradation process in the vacuole while autophagy was active, autophagosomes should accumulate within the vacuole.
Dr Ohsumi cultured mutated yeast lacking vacuolar degradation enzymes and simultaneously stimulated autophagy by starving the cells.
Within hours, the vacuoles were filled with small vesicles that had not been degraded. The vesicles were autophagosomes, and the experiment proved that autophagy exists in yeast cells.
The experiment also provided a method to identify and characterize genes involved in autophagy.
Dr Ohsumi noted that the accumulation of autophagosomes should not occur if genes important for autophagy were inactivated.
So he exposed the yeast cells to a chemical that randomly introduced mutations in many genes, and then he induced autophagy. In this way, he identified 15 genes essential for autophagy in budding yeast.
In his subsequent studies, Dr Ohsumi cloned several of these genes in yeast and mammalian cells and characterized the function of the proteins encoded by these genes.
He found that autophagy is controlled by a cascade of proteins and protein complexes, each regulating a distinct stage of autophagosome initiation and formation.
Insights provided by Dr Ohsumi’s work enabled subsequent research that has revealed the role of autophagy in human physiology and disease.
For more information on Dr Ohsumi and his work, visit the Nobel Prize website. ![]()
Analysis raises concerns about FDA reviewers

Photo by Steven Harbour
Results of an analysis published in The BMJ have raised concerns about how often drug regulators go on to work in the biopharmaceutical industry.
A pair of researchers evaluated 55 medical reviewers who worked at the US Food and Drug Administration (FDA) between 2001 and 2010.
Forty-seven percent of the reviewers left the FDA, and 58% of those who left were subsequently employed by or consulting for the biopharmaceutical industry.
Vinay Prasad, MD, and Jeffrey Bien, MD, both of Oregon Health and Science University in Portland, conducted this research.
The pair began by identifying hematology-oncology drugs approved by the FDA from 2006 to 2010. They then used the FDA database ([email protected]) to compile a list of all medical reviewers for these drugs.
The researchers then searched publicly available information from the Department of Health and Human Services, LinkedIn, and PubMed to determine the reviewers’ subsequent jobs.
The pair identified 55 unique hematology-oncology medical reviewers who reviewed drug applications between 2001 and 2010.
Forty-nine percent (n=27) of these reviewers continue to work exclusively at the FDA. Four percent (n=2) still work at the FDA but hold secondary appointments (1 with a non-FDA-related consulting position and 1 with another government position).
Forty-seven percent (n=26) of the reviewers left the FDA. The researchers were unable to determine subsequent jobs for 14% (n=8) of the reviewers.
However, 27% (n=15) went on to biopharmaceutical industry employment or consulting. Four percent (n=2) went on to other government positions, and 2% (n=1) went on to work in academia.
Drs Prasad and Bien said this analysis is the first to document the rate of the “revolving door” between the FDA and the biopharmaceutical industry, and the results suggest a sizable percentage of FDA medical reviewers who leave the agency subsequently work in the industry.
The researchers said they are concerned by these findings, and they noted that this analysis may have underestimated the extent of the “revolving door” because they could not identify subsequent jobs for all of the reviewers studied. ![]()

Photo by Steven Harbour
Results of an analysis published in The BMJ have raised concerns about how often drug regulators go on to work in the biopharmaceutical industry.
A pair of researchers evaluated 55 medical reviewers who worked at the US Food and Drug Administration (FDA) between 2001 and 2010.
Forty-seven percent of the reviewers left the FDA, and 58% of those who left were subsequently employed by or consulting for the biopharmaceutical industry.
Vinay Prasad, MD, and Jeffrey Bien, MD, both of Oregon Health and Science University in Portland, conducted this research.
The pair began by identifying hematology-oncology drugs approved by the FDA from 2006 to 2010. They then used the FDA database ([email protected]) to compile a list of all medical reviewers for these drugs.
The researchers then searched publicly available information from the Department of Health and Human Services, LinkedIn, and PubMed to determine the reviewers’ subsequent jobs.
The pair identified 55 unique hematology-oncology medical reviewers who reviewed drug applications between 2001 and 2010.
Forty-nine percent (n=27) of these reviewers continue to work exclusively at the FDA. Four percent (n=2) still work at the FDA but hold secondary appointments (1 with a non-FDA-related consulting position and 1 with another government position).
Forty-seven percent (n=26) of the reviewers left the FDA. The researchers were unable to determine subsequent jobs for 14% (n=8) of the reviewers.
However, 27% (n=15) went on to biopharmaceutical industry employment or consulting. Four percent (n=2) went on to other government positions, and 2% (n=1) went on to work in academia.
Drs Prasad and Bien said this analysis is the first to document the rate of the “revolving door” between the FDA and the biopharmaceutical industry, and the results suggest a sizable percentage of FDA medical reviewers who leave the agency subsequently work in the industry.
The researchers said they are concerned by these findings, and they noted that this analysis may have underestimated the extent of the “revolving door” because they could not identify subsequent jobs for all of the reviewers studied. ![]()

Photo by Steven Harbour
Results of an analysis published in The BMJ have raised concerns about how often drug regulators go on to work in the biopharmaceutical industry.
A pair of researchers evaluated 55 medical reviewers who worked at the US Food and Drug Administration (FDA) between 2001 and 2010.
Forty-seven percent of the reviewers left the FDA, and 58% of those who left were subsequently employed by or consulting for the biopharmaceutical industry.
Vinay Prasad, MD, and Jeffrey Bien, MD, both of Oregon Health and Science University in Portland, conducted this research.
The pair began by identifying hematology-oncology drugs approved by the FDA from 2006 to 2010. They then used the FDA database ([email protected]) to compile a list of all medical reviewers for these drugs.
The researchers then searched publicly available information from the Department of Health and Human Services, LinkedIn, and PubMed to determine the reviewers’ subsequent jobs.
The pair identified 55 unique hematology-oncology medical reviewers who reviewed drug applications between 2001 and 2010.
Forty-nine percent (n=27) of these reviewers continue to work exclusively at the FDA. Four percent (n=2) still work at the FDA but hold secondary appointments (1 with a non-FDA-related consulting position and 1 with another government position).
Forty-seven percent (n=26) of the reviewers left the FDA. The researchers were unable to determine subsequent jobs for 14% (n=8) of the reviewers.
However, 27% (n=15) went on to biopharmaceutical industry employment or consulting. Four percent (n=2) went on to other government positions, and 2% (n=1) went on to work in academia.
Drs Prasad and Bien said this analysis is the first to document the rate of the “revolving door” between the FDA and the biopharmaceutical industry, and the results suggest a sizable percentage of FDA medical reviewers who leave the agency subsequently work in the industry.
The researchers said they are concerned by these findings, and they noted that this analysis may have underestimated the extent of the “revolving door” because they could not identify subsequent jobs for all of the reviewers studied. ![]()
Fungus makes mosquitoes more susceptible to malaria
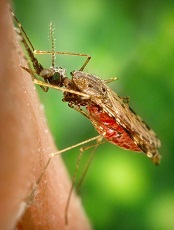
Photo by James Gathany
Researchers say they have identified a fungus that compromises mosquitoes’ immune systems, making them more susceptible to infection with malaria parasites.
Malaria researchers have, in the past, identified microbes that prevent the Anopheles mosquito from being infected by malaria parasites, but this is the first time they have found a microorganism that appears to make the mosquito more likely to become infected with—and then spread—malaria.
The finding was published in Scientific Reports.
“This very common, naturally occurring fungus may have a significant impact on malaria transmission,” said study author George Dimopoulos, PhD, of the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland.
“It doesn’t kill the mosquitoes. It doesn’t make them sick. It just makes them more likely to become infected and thereby to spread the disease. While this fungus is unlikely to be helpful as part of a malaria control strategy, our finding significantly advances our knowledge of the different factors that influence the transmission of malaria.”
For this study, Dr Dimopoulos and his colleagues isolated the Penicillium chrysogenum fungus from the gut of field-caught Anopheles mosquitoes. The team found this fungus made the mosquitoes more susceptible to being infected by Plasmodium parasites through a secreted heat-stable factor.
The researchers said the mechanism behind this increased susceptibility involves upregulation of the mosquitoes’ ornithine decarboxylase gene, which sequesters arginine for polyamine biosynthesis.
They noted that arginine plays an important role in the mosquitoes’ anti-Plasmodium defense as a substrate of nitric oxide production, so the availability of arginine has a direct impact on susceptibility to infection with Plasmodium parasites.
Dr Dimopoulos said Penicillium chrysogenum had not previously been studied in terms of mosquito biology, and he and his team had hoped the fungus would act like several other bacteria that researchers have identified, which prevent mosquitoes from becoming infected with malaria parasites.
Even though Penicillium chrysogenum actually appears to worsen infections, the team believes the fungus can still help researchers in their fight against malaria.
“We have questions we hope this finding will help us to answer, including, ‘Why do we have increased transmission of malaria in some areas and not others when the presence of mosquitoes is the same?’” Dr Dimopoulos said. “This gives us another piece of the complicated malaria puzzle.”
Because environmental microorganisms can vary greatly from region to region, Dr Dimopoulos and his colleagues believe their finding may help explain variations in the prevalence of malaria in different geographic areas. ![]()

Photo by James Gathany
Researchers say they have identified a fungus that compromises mosquitoes’ immune systems, making them more susceptible to infection with malaria parasites.
Malaria researchers have, in the past, identified microbes that prevent the Anopheles mosquito from being infected by malaria parasites, but this is the first time they have found a microorganism that appears to make the mosquito more likely to become infected with—and then spread—malaria.
The finding was published in Scientific Reports.
“This very common, naturally occurring fungus may have a significant impact on malaria transmission,” said study author George Dimopoulos, PhD, of the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland.
“It doesn’t kill the mosquitoes. It doesn’t make them sick. It just makes them more likely to become infected and thereby to spread the disease. While this fungus is unlikely to be helpful as part of a malaria control strategy, our finding significantly advances our knowledge of the different factors that influence the transmission of malaria.”
For this study, Dr Dimopoulos and his colleagues isolated the Penicillium chrysogenum fungus from the gut of field-caught Anopheles mosquitoes. The team found this fungus made the mosquitoes more susceptible to being infected by Plasmodium parasites through a secreted heat-stable factor.
The researchers said the mechanism behind this increased susceptibility involves upregulation of the mosquitoes’ ornithine decarboxylase gene, which sequesters arginine for polyamine biosynthesis.
They noted that arginine plays an important role in the mosquitoes’ anti-Plasmodium defense as a substrate of nitric oxide production, so the availability of arginine has a direct impact on susceptibility to infection with Plasmodium parasites.
Dr Dimopoulos said Penicillium chrysogenum had not previously been studied in terms of mosquito biology, and he and his team had hoped the fungus would act like several other bacteria that researchers have identified, which prevent mosquitoes from becoming infected with malaria parasites.
Even though Penicillium chrysogenum actually appears to worsen infections, the team believes the fungus can still help researchers in their fight against malaria.
“We have questions we hope this finding will help us to answer, including, ‘Why do we have increased transmission of malaria in some areas and not others when the presence of mosquitoes is the same?’” Dr Dimopoulos said. “This gives us another piece of the complicated malaria puzzle.”
Because environmental microorganisms can vary greatly from region to region, Dr Dimopoulos and his colleagues believe their finding may help explain variations in the prevalence of malaria in different geographic areas. ![]()

Photo by James Gathany
Researchers say they have identified a fungus that compromises mosquitoes’ immune systems, making them more susceptible to infection with malaria parasites.
Malaria researchers have, in the past, identified microbes that prevent the Anopheles mosquito from being infected by malaria parasites, but this is the first time they have found a microorganism that appears to make the mosquito more likely to become infected with—and then spread—malaria.
The finding was published in Scientific Reports.
“This very common, naturally occurring fungus may have a significant impact on malaria transmission,” said study author George Dimopoulos, PhD, of the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland.
“It doesn’t kill the mosquitoes. It doesn’t make them sick. It just makes them more likely to become infected and thereby to spread the disease. While this fungus is unlikely to be helpful as part of a malaria control strategy, our finding significantly advances our knowledge of the different factors that influence the transmission of malaria.”
For this study, Dr Dimopoulos and his colleagues isolated the Penicillium chrysogenum fungus from the gut of field-caught Anopheles mosquitoes. The team found this fungus made the mosquitoes more susceptible to being infected by Plasmodium parasites through a secreted heat-stable factor.
The researchers said the mechanism behind this increased susceptibility involves upregulation of the mosquitoes’ ornithine decarboxylase gene, which sequesters arginine for polyamine biosynthesis.
They noted that arginine plays an important role in the mosquitoes’ anti-Plasmodium defense as a substrate of nitric oxide production, so the availability of arginine has a direct impact on susceptibility to infection with Plasmodium parasites.
Dr Dimopoulos said Penicillium chrysogenum had not previously been studied in terms of mosquito biology, and he and his team had hoped the fungus would act like several other bacteria that researchers have identified, which prevent mosquitoes from becoming infected with malaria parasites.
Even though Penicillium chrysogenum actually appears to worsen infections, the team believes the fungus can still help researchers in their fight against malaria.
“We have questions we hope this finding will help us to answer, including, ‘Why do we have increased transmission of malaria in some areas and not others when the presence of mosquitoes is the same?’” Dr Dimopoulos said. “This gives us another piece of the complicated malaria puzzle.”
Because environmental microorganisms can vary greatly from region to region, Dr Dimopoulos and his colleagues believe their finding may help explain variations in the prevalence of malaria in different geographic areas.
Study shows RT underused in developing countries

Photo courtesy of ASTRO
BOSTON—A new study suggests that roughly half of cancer patients in developing countries need radiation therapy (RT) to treat their disease, but many of these patients do not have access to it.
Examining 9 developing countries, investigators found that between 18% and 82% of patients who can benefit from RT do not receive the treatment.
These findings were presented at ASTRO’s 58th Annual Meeting (abstract 82).
“Access to radiation therapy remains limited in low-and middle-income countries,” said study investigator Elena Fidarova, MD, of the International Atomic Energy Agency in Vienna, Austria.
“In Ghana and the Philippines, for example, about 8 in 10 cancer patients who need radiation therapy will not receive needed treatment.”
Dr Fidarova and her colleagues conducted this study to assess levels of optimal and actual RT utilization (RTU) and calculate unmet RT need in 9 developing countries—Costa Rica, Ghana, Malaysia, the Philippines, Romania, Serbia, Slovenia, Tunisia, and Uruguay.
The investigators determined the optimal and actual RTU rates for each country. The optimal RTU rate is the proportion of all newly diagnosed cancer patients who have an indication for RT at least once in their lifetime.
An indication for RT was defined as a clinical scenario for which RT is recommended as the treatment of choice because there is evidence of its superiority to alternative modalities and/or no treatment (eg, better survival, local control, or quality of life profiles).
In clinical situations where RT was equivalent to other treatment options, all comparable modalities were included in the model, and a subsequent sensitivity analysis was conducted to determine the proportion of these patients for whom RT was indicated.
Results
The median optimal RTU for all countries was 52%. Optimal RTU rates ranged from a low of 47% for Costa Rica to a high of 56% for Tunisia. Differences in optimal RTU rates are attributable to varying incidence rates of cancer types in each country.
The median actual RTU rate was roughly half of optimal utilization, suggesting that nearly half of cancer patients across these 9 countries combined may not be receiving adequate care for their disease.
The median actual RTU rate was 28%. The lowest rates of utilization were in Ghana (9%) and the Philippines (10.3%), while the highest utilization rates were in Tunisia (46%) and Uruguay (37%).
Actual RTU rates were lower than optimal RTU rates for all 9 countries, with the smallest difference in Tunisia and the widest gap in Ghana—at nearly 43 percentage points.
The median level of unmet need was 47% for all countries combined.
Ghana and the Philippines had the highest levels of unmet need, at 82.3% and 80.5%, respectively. Costa Rica and Tunisia had the lowest levels of unmet need, at 25.5% and 18%, respectively.
The unmet need was especially high in countries with limited resources and a large population. The number of teletherapy machines per 1000 cancer cases ranged from a high of 1.3 in Tunisia to a low of 0.19 in Ghana.
The strong correlation between the actual RTU rates and the number of teletherapy machines per 1000 cancer cases/year in each country confirms that, although other access factors may be at play, the availability of RT machines is an important factor in RT utilization.
“Differences between optimal and actual RTU rates and the high percentage of unmet RT need likely stem from a number of complex reasons, although inadequate capacity for radiation therapy is the most obvious factor,” Dr Fidarova said.
“As obstacles in access to existing RT services—such as inadequate referral patterns, affordability of treatment, and geographical distribution of centers—differ by country, so does the ideal mix of solutions.”

Photo courtesy of ASTRO
BOSTON—A new study suggests that roughly half of cancer patients in developing countries need radiation therapy (RT) to treat their disease, but many of these patients do not have access to it.
Examining 9 developing countries, investigators found that between 18% and 82% of patients who can benefit from RT do not receive the treatment.
These findings were presented at ASTRO’s 58th Annual Meeting (abstract 82).
“Access to radiation therapy remains limited in low-and middle-income countries,” said study investigator Elena Fidarova, MD, of the International Atomic Energy Agency in Vienna, Austria.
“In Ghana and the Philippines, for example, about 8 in 10 cancer patients who need radiation therapy will not receive needed treatment.”
Dr Fidarova and her colleagues conducted this study to assess levels of optimal and actual RT utilization (RTU) and calculate unmet RT need in 9 developing countries—Costa Rica, Ghana, Malaysia, the Philippines, Romania, Serbia, Slovenia, Tunisia, and Uruguay.
The investigators determined the optimal and actual RTU rates for each country. The optimal RTU rate is the proportion of all newly diagnosed cancer patients who have an indication for RT at least once in their lifetime.
An indication for RT was defined as a clinical scenario for which RT is recommended as the treatment of choice because there is evidence of its superiority to alternative modalities and/or no treatment (eg, better survival, local control, or quality of life profiles).
In clinical situations where RT was equivalent to other treatment options, all comparable modalities were included in the model, and a subsequent sensitivity analysis was conducted to determine the proportion of these patients for whom RT was indicated.
Results
The median optimal RTU for all countries was 52%. Optimal RTU rates ranged from a low of 47% for Costa Rica to a high of 56% for Tunisia. Differences in optimal RTU rates are attributable to varying incidence rates of cancer types in each country.
The median actual RTU rate was roughly half of optimal utilization, suggesting that nearly half of cancer patients across these 9 countries combined may not be receiving adequate care for their disease.
The median actual RTU rate was 28%. The lowest rates of utilization were in Ghana (9%) and the Philippines (10.3%), while the highest utilization rates were in Tunisia (46%) and Uruguay (37%).
Actual RTU rates were lower than optimal RTU rates for all 9 countries, with the smallest difference in Tunisia and the widest gap in Ghana—at nearly 43 percentage points.
The median level of unmet need was 47% for all countries combined.
Ghana and the Philippines had the highest levels of unmet need, at 82.3% and 80.5%, respectively. Costa Rica and Tunisia had the lowest levels of unmet need, at 25.5% and 18%, respectively.
The unmet need was especially high in countries with limited resources and a large population. The number of teletherapy machines per 1000 cancer cases ranged from a high of 1.3 in Tunisia to a low of 0.19 in Ghana.
The strong correlation between the actual RTU rates and the number of teletherapy machines per 1000 cancer cases/year in each country confirms that, although other access factors may be at play, the availability of RT machines is an important factor in RT utilization.
“Differences between optimal and actual RTU rates and the high percentage of unmet RT need likely stem from a number of complex reasons, although inadequate capacity for radiation therapy is the most obvious factor,” Dr Fidarova said.
“As obstacles in access to existing RT services—such as inadequate referral patterns, affordability of treatment, and geographical distribution of centers—differ by country, so does the ideal mix of solutions.”

Photo courtesy of ASTRO
BOSTON—A new study suggests that roughly half of cancer patients in developing countries need radiation therapy (RT) to treat their disease, but many of these patients do not have access to it.
Examining 9 developing countries, investigators found that between 18% and 82% of patients who can benefit from RT do not receive the treatment.
These findings were presented at ASTRO’s 58th Annual Meeting (abstract 82).
“Access to radiation therapy remains limited in low-and middle-income countries,” said study investigator Elena Fidarova, MD, of the International Atomic Energy Agency in Vienna, Austria.
“In Ghana and the Philippines, for example, about 8 in 10 cancer patients who need radiation therapy will not receive needed treatment.”
Dr Fidarova and her colleagues conducted this study to assess levels of optimal and actual RT utilization (RTU) and calculate unmet RT need in 9 developing countries—Costa Rica, Ghana, Malaysia, the Philippines, Romania, Serbia, Slovenia, Tunisia, and Uruguay.
The investigators determined the optimal and actual RTU rates for each country. The optimal RTU rate is the proportion of all newly diagnosed cancer patients who have an indication for RT at least once in their lifetime.
An indication for RT was defined as a clinical scenario for which RT is recommended as the treatment of choice because there is evidence of its superiority to alternative modalities and/or no treatment (eg, better survival, local control, or quality of life profiles).
In clinical situations where RT was equivalent to other treatment options, all comparable modalities were included in the model, and a subsequent sensitivity analysis was conducted to determine the proportion of these patients for whom RT was indicated.
Results
The median optimal RTU for all countries was 52%. Optimal RTU rates ranged from a low of 47% for Costa Rica to a high of 56% for Tunisia. Differences in optimal RTU rates are attributable to varying incidence rates of cancer types in each country.
The median actual RTU rate was roughly half of optimal utilization, suggesting that nearly half of cancer patients across these 9 countries combined may not be receiving adequate care for their disease.
The median actual RTU rate was 28%. The lowest rates of utilization were in Ghana (9%) and the Philippines (10.3%), while the highest utilization rates were in Tunisia (46%) and Uruguay (37%).
Actual RTU rates were lower than optimal RTU rates for all 9 countries, with the smallest difference in Tunisia and the widest gap in Ghana—at nearly 43 percentage points.
The median level of unmet need was 47% for all countries combined.
Ghana and the Philippines had the highest levels of unmet need, at 82.3% and 80.5%, respectively. Costa Rica and Tunisia had the lowest levels of unmet need, at 25.5% and 18%, respectively.
The unmet need was especially high in countries with limited resources and a large population. The number of teletherapy machines per 1000 cancer cases ranged from a high of 1.3 in Tunisia to a low of 0.19 in Ghana.
The strong correlation between the actual RTU rates and the number of teletherapy machines per 1000 cancer cases/year in each country confirms that, although other access factors may be at play, the availability of RT machines is an important factor in RT utilization.
“Differences between optimal and actual RTU rates and the high percentage of unmet RT need likely stem from a number of complex reasons, although inadequate capacity for radiation therapy is the most obvious factor,” Dr Fidarova said.
“As obstacles in access to existing RT services—such as inadequate referral patterns, affordability of treatment, and geographical distribution of centers—differ by country, so does the ideal mix of solutions.”
OSC calls for further review of allegations about Zika test

Photo by Graham Colm
The US Office of Special Counsel (OSC) has called for further review of allegations made about the Trioplex assay, a test used to detect Zika and other viruses.
A whistleblower recently alleged that the Centers for Disease Control and Prevention (CDC) has been using and promoting the Trioplex assay even though this test is nearly 40% less effective for Zika virus detection than another test, the Singleplex assay.
The CDC conducted an investigation that suggested this claim is not accurate, but the OSC has recommended additional review of the issue. (The OSC is an independent federal investigative and prosecutorial agency.)
Allegations
The allegations about the Trioplex assay were made by Robert Lanciotti, PhD, a CDC microbiologist based in Fort Collins, Colorado.
Dr Lanciotti conducted a study in which the Trioplex assay—which tests for Zika, dengue, and chikungunya—missed 39% of Zika infections detected by the Singleplex assay, which only tests for Zika.
Dr Lanciotti also raised concerns that the CDC’s Emergency Operations Center (EOC) withheld from public health laboratories information about the sensitivity differences between the Trioplex and Singleplex assays.
He said the CDC may have given laboratories the mistaken impression that Trioplex was a better test.
The Singleplex assay was made by Dr Lanciotti’s lab, while the Trioplex assay was developed at the CDC’s dengue branch lab in Puerto Rico.
Investigation
The OSC referred Dr Lanciotti’s claims to the Department of Health and Human Services (HHS) for investigation on July 1, 2016.
HHS Secretary Sylvia Mathews Burwell directed Steve Monroe, PhD, associate director for laboratory science and safety at the CDC, to conduct the investigation. The investigative team did not include employees who worked in EOC or in the zoonotic infectious diseases branch of the CDC.
The CDC said its investigation did not substantiate Dr Lanciotti’s claims. Investigators said they were unable to reach a “statistically valid conclusion about the relative performance” of the tests.
The CDC’s report pointed to a study conducted by its dengue branch in Puerto Rico that “found no difference in sensitivity” between the assays. The Trioplex assay was developed at this lab.
The report also said the CDC acted reasonably when it withheld information about the sensitivity differences between the Trioplex and Singleplex assays because there was conflicting data from different labs. The investigators said releasing that data could have created “considerable confusion during an ongoing emergency response.”
The CDC also noted that, in late August, the agency made changes intended to improve the sensitivity of the Trioplex assay, such as increasing sample volumes and allowing whole blood as a specimen type.
Response
Dr Lanciotti took issue with several points in the CDC’s report. Perhaps most importantly, he said “there was clearly enough data to warrant a ‘pause’ in the recommendation of the Trioplex until an extensive comparison could be performed.”
He referenced a multicenter study conducted independently by the Blood Systems Research Institute in San Francisco, California, which demonstrated the Trioplex assay’s lower sensitivity.
Dr Lanciotti added that the method used by this institution to assess the tests is “the most accurate method to evaluate the clinical sensitivity . . . of individual assays.”
Reassignment
Prior to disclosing his concerns to the OSC, Dr Lanciotti voiced the concerns internally and in an email to state public health officials in April 2016.
In May, he was reassigned to a non‐supervisory position within his lab. After the reassignment, Dr Lanciotti filed a whistleblower retaliation claim alleging that his diminished duties, from lab chief to a non‐supervisory position, was in reprisal for his disclosures.
After an investigation, the OSC secured an agreement from the CDC to reinstate Dr Lanciotti as chief of his lab.
OSC assessment
In a letter to President Barack Obama, Carolyn Lerner, head of the OSC, said the CDC “conducted a thorough investigation into Dr Lanciotti’s allegations, and its findings appear reasonable.”
On the other hand, Dr Lanciotti has raised “serious concerns” about the CDC’s findings.
“As the agency contemplates additional improvements or changes to the Zika testing protocol, I encourage CDC to review Dr Lanciotti’s comments, respond to each of his concerns, and utilize his expertise as the agency works to ensure it is implementing the most effective testing methods in response to this public health emergency,” Lerner said.
“I also encourage the CDC to promote scientific debate within its labs. Whistleblowers should be encouraged to speak out on matters of public concern.”

Photo by Graham Colm
The US Office of Special Counsel (OSC) has called for further review of allegations made about the Trioplex assay, a test used to detect Zika and other viruses.
A whistleblower recently alleged that the Centers for Disease Control and Prevention (CDC) has been using and promoting the Trioplex assay even though this test is nearly 40% less effective for Zika virus detection than another test, the Singleplex assay.
The CDC conducted an investigation that suggested this claim is not accurate, but the OSC has recommended additional review of the issue. (The OSC is an independent federal investigative and prosecutorial agency.)
Allegations
The allegations about the Trioplex assay were made by Robert Lanciotti, PhD, a CDC microbiologist based in Fort Collins, Colorado.
Dr Lanciotti conducted a study in which the Trioplex assay—which tests for Zika, dengue, and chikungunya—missed 39% of Zika infections detected by the Singleplex assay, which only tests for Zika.
Dr Lanciotti also raised concerns that the CDC’s Emergency Operations Center (EOC) withheld from public health laboratories information about the sensitivity differences between the Trioplex and Singleplex assays.
He said the CDC may have given laboratories the mistaken impression that Trioplex was a better test.
The Singleplex assay was made by Dr Lanciotti’s lab, while the Trioplex assay was developed at the CDC’s dengue branch lab in Puerto Rico.
Investigation
The OSC referred Dr Lanciotti’s claims to the Department of Health and Human Services (HHS) for investigation on July 1, 2016.
HHS Secretary Sylvia Mathews Burwell directed Steve Monroe, PhD, associate director for laboratory science and safety at the CDC, to conduct the investigation. The investigative team did not include employees who worked in EOC or in the zoonotic infectious diseases branch of the CDC.
The CDC said its investigation did not substantiate Dr Lanciotti’s claims. Investigators said they were unable to reach a “statistically valid conclusion about the relative performance” of the tests.
The CDC’s report pointed to a study conducted by its dengue branch in Puerto Rico that “found no difference in sensitivity” between the assays. The Trioplex assay was developed at this lab.
The report also said the CDC acted reasonably when it withheld information about the sensitivity differences between the Trioplex and Singleplex assays because there was conflicting data from different labs. The investigators said releasing that data could have created “considerable confusion during an ongoing emergency response.”
The CDC also noted that, in late August, the agency made changes intended to improve the sensitivity of the Trioplex assay, such as increasing sample volumes and allowing whole blood as a specimen type.
Response
Dr Lanciotti took issue with several points in the CDC’s report. Perhaps most importantly, he said “there was clearly enough data to warrant a ‘pause’ in the recommendation of the Trioplex until an extensive comparison could be performed.”
He referenced a multicenter study conducted independently by the Blood Systems Research Institute in San Francisco, California, which demonstrated the Trioplex assay’s lower sensitivity.
Dr Lanciotti added that the method used by this institution to assess the tests is “the most accurate method to evaluate the clinical sensitivity . . . of individual assays.”
Reassignment
Prior to disclosing his concerns to the OSC, Dr Lanciotti voiced the concerns internally and in an email to state public health officials in April 2016.
In May, he was reassigned to a non‐supervisory position within his lab. After the reassignment, Dr Lanciotti filed a whistleblower retaliation claim alleging that his diminished duties, from lab chief to a non‐supervisory position, was in reprisal for his disclosures.
After an investigation, the OSC secured an agreement from the CDC to reinstate Dr Lanciotti as chief of his lab.
OSC assessment
In a letter to President Barack Obama, Carolyn Lerner, head of the OSC, said the CDC “conducted a thorough investigation into Dr Lanciotti’s allegations, and its findings appear reasonable.”
On the other hand, Dr Lanciotti has raised “serious concerns” about the CDC’s findings.
“As the agency contemplates additional improvements or changes to the Zika testing protocol, I encourage CDC to review Dr Lanciotti’s comments, respond to each of his concerns, and utilize his expertise as the agency works to ensure it is implementing the most effective testing methods in response to this public health emergency,” Lerner said.
“I also encourage the CDC to promote scientific debate within its labs. Whistleblowers should be encouraged to speak out on matters of public concern.”

Photo by Graham Colm
The US Office of Special Counsel (OSC) has called for further review of allegations made about the Trioplex assay, a test used to detect Zika and other viruses.
A whistleblower recently alleged that the Centers for Disease Control and Prevention (CDC) has been using and promoting the Trioplex assay even though this test is nearly 40% less effective for Zika virus detection than another test, the Singleplex assay.
The CDC conducted an investigation that suggested this claim is not accurate, but the OSC has recommended additional review of the issue. (The OSC is an independent federal investigative and prosecutorial agency.)
Allegations
The allegations about the Trioplex assay were made by Robert Lanciotti, PhD, a CDC microbiologist based in Fort Collins, Colorado.
Dr Lanciotti conducted a study in which the Trioplex assay—which tests for Zika, dengue, and chikungunya—missed 39% of Zika infections detected by the Singleplex assay, which only tests for Zika.
Dr Lanciotti also raised concerns that the CDC’s Emergency Operations Center (EOC) withheld from public health laboratories information about the sensitivity differences between the Trioplex and Singleplex assays.
He said the CDC may have given laboratories the mistaken impression that Trioplex was a better test.
The Singleplex assay was made by Dr Lanciotti’s lab, while the Trioplex assay was developed at the CDC’s dengue branch lab in Puerto Rico.
Investigation
The OSC referred Dr Lanciotti’s claims to the Department of Health and Human Services (HHS) for investigation on July 1, 2016.
HHS Secretary Sylvia Mathews Burwell directed Steve Monroe, PhD, associate director for laboratory science and safety at the CDC, to conduct the investigation. The investigative team did not include employees who worked in EOC or in the zoonotic infectious diseases branch of the CDC.
The CDC said its investigation did not substantiate Dr Lanciotti’s claims. Investigators said they were unable to reach a “statistically valid conclusion about the relative performance” of the tests.
The CDC’s report pointed to a study conducted by its dengue branch in Puerto Rico that “found no difference in sensitivity” between the assays. The Trioplex assay was developed at this lab.
The report also said the CDC acted reasonably when it withheld information about the sensitivity differences between the Trioplex and Singleplex assays because there was conflicting data from different labs. The investigators said releasing that data could have created “considerable confusion during an ongoing emergency response.”
The CDC also noted that, in late August, the agency made changes intended to improve the sensitivity of the Trioplex assay, such as increasing sample volumes and allowing whole blood as a specimen type.
Response
Dr Lanciotti took issue with several points in the CDC’s report. Perhaps most importantly, he said “there was clearly enough data to warrant a ‘pause’ in the recommendation of the Trioplex until an extensive comparison could be performed.”
He referenced a multicenter study conducted independently by the Blood Systems Research Institute in San Francisco, California, which demonstrated the Trioplex assay’s lower sensitivity.
Dr Lanciotti added that the method used by this institution to assess the tests is “the most accurate method to evaluate the clinical sensitivity . . . of individual assays.”
Reassignment
Prior to disclosing his concerns to the OSC, Dr Lanciotti voiced the concerns internally and in an email to state public health officials in April 2016.
In May, he was reassigned to a non‐supervisory position within his lab. After the reassignment, Dr Lanciotti filed a whistleblower retaliation claim alleging that his diminished duties, from lab chief to a non‐supervisory position, was in reprisal for his disclosures.
After an investigation, the OSC secured an agreement from the CDC to reinstate Dr Lanciotti as chief of his lab.
OSC assessment
In a letter to President Barack Obama, Carolyn Lerner, head of the OSC, said the CDC “conducted a thorough investigation into Dr Lanciotti’s allegations, and its findings appear reasonable.”
On the other hand, Dr Lanciotti has raised “serious concerns” about the CDC’s findings.
“As the agency contemplates additional improvements or changes to the Zika testing protocol, I encourage CDC to review Dr Lanciotti’s comments, respond to each of his concerns, and utilize his expertise as the agency works to ensure it is implementing the most effective testing methods in response to this public health emergency,” Lerner said.
“I also encourage the CDC to promote scientific debate within its labs. Whistleblowers should be encouraged to speak out on matters of public concern.”
Database of research regulations gets update

for a clinical trial
Photo by Esther Dyson
The ClinRegs website has been updated and upgraded, according to the National Institute of Allergy and Infectious Diseases.
ClinRegs is an online database of country-specific information on clinical research regulations that was launched in 2014.
Now, the website houses regulatory information for 17 countries and has an interactive map on its homepage to provide a clearer picture of the countries included.
ClinRegs also has a hyperlinked table of contents on each country page that is intended to provide easier navigation.
Drop-down menus allow users to switch between country profiles and make comparisons between countries.
In addition, ClinRegs now provides a Quick Facts table with discrete pieces of information for each country.
And a feedback link has been added to the site to make it easy for users to submit comments or updates to regulations.

for a clinical trial
Photo by Esther Dyson
The ClinRegs website has been updated and upgraded, according to the National Institute of Allergy and Infectious Diseases.
ClinRegs is an online database of country-specific information on clinical research regulations that was launched in 2014.
Now, the website houses regulatory information for 17 countries and has an interactive map on its homepage to provide a clearer picture of the countries included.
ClinRegs also has a hyperlinked table of contents on each country page that is intended to provide easier navigation.
Drop-down menus allow users to switch between country profiles and make comparisons between countries.
In addition, ClinRegs now provides a Quick Facts table with discrete pieces of information for each country.
And a feedback link has been added to the site to make it easy for users to submit comments or updates to regulations.

for a clinical trial
Photo by Esther Dyson
The ClinRegs website has been updated and upgraded, according to the National Institute of Allergy and Infectious Diseases.
ClinRegs is an online database of country-specific information on clinical research regulations that was launched in 2014.
Now, the website houses regulatory information for 17 countries and has an interactive map on its homepage to provide a clearer picture of the countries included.
ClinRegs also has a hyperlinked table of contents on each country page that is intended to provide easier navigation.
Drop-down menus allow users to switch between country profiles and make comparisons between countries.
In addition, ClinRegs now provides a Quick Facts table with discrete pieces of information for each country.
And a feedback link has been added to the site to make it easy for users to submit comments or updates to regulations.