User login
Tool provides info for cancer patients, survivors
receiving treatment
Photo by Rhoda Baer
The American Cancer Society and National Cancer Institute have launched an online tool for cancer patients and survivors.
The tool, Springboard Beyond Cancer, was designed to help these individuals address medical, psychosocial, and wellness needs during and after treatment.
Springboard Beyond Cancer provides information to help cancer patients and survivors manage ongoing cancer-related symptoms, deal with stress, ensure healthy behavior, communicate better with healthcare teams, and seek support from friends and family.
“With Springboard Beyond Cancer, we want to empower cancer survivors by giving them the information they need to help identify issues, set goals, and create a plan to more smoothly navigate the cancer journey and take control of their health,” said Corinne Leach, PhD, a behavioral scientist and strategic director in the Behavioral Research Center at the American Cancer Society.
“We hope that Springboard Beyond Cancer, along with the close collaboration of their medical team, can help cancer survivors reduce their disease burden and improve their overall wellbeing,” added Erik Augustson, PhD, program director at the National Cancer Institute. ![]()

receiving treatment
Photo by Rhoda Baer
The American Cancer Society and National Cancer Institute have launched an online tool for cancer patients and survivors.
The tool, Springboard Beyond Cancer, was designed to help these individuals address medical, psychosocial, and wellness needs during and after treatment.
Springboard Beyond Cancer provides information to help cancer patients and survivors manage ongoing cancer-related symptoms, deal with stress, ensure healthy behavior, communicate better with healthcare teams, and seek support from friends and family.
“With Springboard Beyond Cancer, we want to empower cancer survivors by giving them the information they need to help identify issues, set goals, and create a plan to more smoothly navigate the cancer journey and take control of their health,” said Corinne Leach, PhD, a behavioral scientist and strategic director in the Behavioral Research Center at the American Cancer Society.
“We hope that Springboard Beyond Cancer, along with the close collaboration of their medical team, can help cancer survivors reduce their disease burden and improve their overall wellbeing,” added Erik Augustson, PhD, program director at the National Cancer Institute. ![]()

receiving treatment
Photo by Rhoda Baer
The American Cancer Society and National Cancer Institute have launched an online tool for cancer patients and survivors.
The tool, Springboard Beyond Cancer, was designed to help these individuals address medical, psychosocial, and wellness needs during and after treatment.
Springboard Beyond Cancer provides information to help cancer patients and survivors manage ongoing cancer-related symptoms, deal with stress, ensure healthy behavior, communicate better with healthcare teams, and seek support from friends and family.
“With Springboard Beyond Cancer, we want to empower cancer survivors by giving them the information they need to help identify issues, set goals, and create a plan to more smoothly navigate the cancer journey and take control of their health,” said Corinne Leach, PhD, a behavioral scientist and strategic director in the Behavioral Research Center at the American Cancer Society.
“We hope that Springboard Beyond Cancer, along with the close collaboration of their medical team, can help cancer survivors reduce their disease burden and improve their overall wellbeing,” added Erik Augustson, PhD, program director at the National Cancer Institute. ![]()
Link between hemolysis and infection explained

plate showing a positive
streptococcus infection
Photo by Bill Branson
Patients who suffer from hemolysis have an increased risk of developing bacterial infections, and new research provides an explanation for this phenomenon.
The study refutes the idea that excess circulating iron is to blame.
Instead, it suggests that heme prevents macrophages from engulfing bacteria. And targeting this activity might reduce the risk of bacterial infection in patients with hemolytic disorders.
Sylvia Knapp, MD, PhD, of the Medical University of Vienna in Austria, and her colleagues reported these findings in Nature Immunology.
For decades, iron has been considered the prime suspect responsible for the high rate of bacterial infections in patients with hemolysis. Iron has long been established as an essential nutrient for bacteria.
Since hemolysis leads to the release of iron-containing heme, the threat of serious bacterial infections in these patients was attributed to the excess availability of circulating iron (heme).
However, Dr Knapp and her colleagues found that heme does not act as a willing nutrient to bacteria.
“Using in vitro and preclinical models, we could clearly demonstrate that heme-derived iron is not at all vital for bacterial growth,” said Rui Martins, a PhD student at the Medical University of Vienna.
“In contrast, we found that heme acts on macrophages, the most significant immune cells that are required for mounting an antibacterial response, and it furthermore prevented these cells from eliminating bacteria.”
Heme interfered with the cytoskeleton of macrophages, thereby immobilizing them.
“Heme causes cells to form numerous spikes—like hair standing on end—and then ‘stuns’ the cells within minutes,” Martins explained. “It is reminiscent of a cartoon character sticking his finger in an electrical outlet.”
The cytoskeleton is crucial for the basic functions of macrophages. It consists of long, branching filaments that act as the cell’s internal, highly flexible, and mobile framework.
Through targeted build-up and breakdown of these filaments, phagocytes can move in any direction and engulf invading bacteria. However, this requires a finely tuned signaling program in which the protein DOCK8 plays a central role.
“Through chemical proteomics and biochemical experiments, we discovered that heme interacted with DOCK8, which led to the permanent activation of its downstream target, Cdc42, with deleterious effects,” Dr Knapp said.
In the presence of heme, the cytoskeletal resilience was lost, as filaments grew rampant in all directions, resulting in macrophage paralysis. The cells lost their ability to shape-shift and could no longer chase down and engulf the invading bacteria, allowing the bacteria to multiply virtually unrestricted.
However, Dr Knapp and her colleagues found that an antimalarial drug can restore the functionality of these paralyzed macrophages.
“Quinine, which is clinically used to treat malaria and is suspected to bind heme, blocks the interaction of heme with DOCK8 and thereby improves the outcome from sepsis,” Dr Knapp said.
“This is very promising. We conclusively demonstrate that it is indeed feasible to therapeutically ‘protect’ immune cells and to restore the body’s immune defense against bacteria in hemolytic conditions.” ![]()

plate showing a positive
streptococcus infection
Photo by Bill Branson
Patients who suffer from hemolysis have an increased risk of developing bacterial infections, and new research provides an explanation for this phenomenon.
The study refutes the idea that excess circulating iron is to blame.
Instead, it suggests that heme prevents macrophages from engulfing bacteria. And targeting this activity might reduce the risk of bacterial infection in patients with hemolytic disorders.
Sylvia Knapp, MD, PhD, of the Medical University of Vienna in Austria, and her colleagues reported these findings in Nature Immunology.
For decades, iron has been considered the prime suspect responsible for the high rate of bacterial infections in patients with hemolysis. Iron has long been established as an essential nutrient for bacteria.
Since hemolysis leads to the release of iron-containing heme, the threat of serious bacterial infections in these patients was attributed to the excess availability of circulating iron (heme).
However, Dr Knapp and her colleagues found that heme does not act as a willing nutrient to bacteria.
“Using in vitro and preclinical models, we could clearly demonstrate that heme-derived iron is not at all vital for bacterial growth,” said Rui Martins, a PhD student at the Medical University of Vienna.
“In contrast, we found that heme acts on macrophages, the most significant immune cells that are required for mounting an antibacterial response, and it furthermore prevented these cells from eliminating bacteria.”
Heme interfered with the cytoskeleton of macrophages, thereby immobilizing them.
“Heme causes cells to form numerous spikes—like hair standing on end—and then ‘stuns’ the cells within minutes,” Martins explained. “It is reminiscent of a cartoon character sticking his finger in an electrical outlet.”
The cytoskeleton is crucial for the basic functions of macrophages. It consists of long, branching filaments that act as the cell’s internal, highly flexible, and mobile framework.
Through targeted build-up and breakdown of these filaments, phagocytes can move in any direction and engulf invading bacteria. However, this requires a finely tuned signaling program in which the protein DOCK8 plays a central role.
“Through chemical proteomics and biochemical experiments, we discovered that heme interacted with DOCK8, which led to the permanent activation of its downstream target, Cdc42, with deleterious effects,” Dr Knapp said.
In the presence of heme, the cytoskeletal resilience was lost, as filaments grew rampant in all directions, resulting in macrophage paralysis. The cells lost their ability to shape-shift and could no longer chase down and engulf the invading bacteria, allowing the bacteria to multiply virtually unrestricted.
However, Dr Knapp and her colleagues found that an antimalarial drug can restore the functionality of these paralyzed macrophages.
“Quinine, which is clinically used to treat malaria and is suspected to bind heme, blocks the interaction of heme with DOCK8 and thereby improves the outcome from sepsis,” Dr Knapp said.
“This is very promising. We conclusively demonstrate that it is indeed feasible to therapeutically ‘protect’ immune cells and to restore the body’s immune defense against bacteria in hemolytic conditions.” ![]()

plate showing a positive
streptococcus infection
Photo by Bill Branson
Patients who suffer from hemolysis have an increased risk of developing bacterial infections, and new research provides an explanation for this phenomenon.
The study refutes the idea that excess circulating iron is to blame.
Instead, it suggests that heme prevents macrophages from engulfing bacteria. And targeting this activity might reduce the risk of bacterial infection in patients with hemolytic disorders.
Sylvia Knapp, MD, PhD, of the Medical University of Vienna in Austria, and her colleagues reported these findings in Nature Immunology.
For decades, iron has been considered the prime suspect responsible for the high rate of bacterial infections in patients with hemolysis. Iron has long been established as an essential nutrient for bacteria.
Since hemolysis leads to the release of iron-containing heme, the threat of serious bacterial infections in these patients was attributed to the excess availability of circulating iron (heme).
However, Dr Knapp and her colleagues found that heme does not act as a willing nutrient to bacteria.
“Using in vitro and preclinical models, we could clearly demonstrate that heme-derived iron is not at all vital for bacterial growth,” said Rui Martins, a PhD student at the Medical University of Vienna.
“In contrast, we found that heme acts on macrophages, the most significant immune cells that are required for mounting an antibacterial response, and it furthermore prevented these cells from eliminating bacteria.”
Heme interfered with the cytoskeleton of macrophages, thereby immobilizing them.
“Heme causes cells to form numerous spikes—like hair standing on end—and then ‘stuns’ the cells within minutes,” Martins explained. “It is reminiscent of a cartoon character sticking his finger in an electrical outlet.”
The cytoskeleton is crucial for the basic functions of macrophages. It consists of long, branching filaments that act as the cell’s internal, highly flexible, and mobile framework.
Through targeted build-up and breakdown of these filaments, phagocytes can move in any direction and engulf invading bacteria. However, this requires a finely tuned signaling program in which the protein DOCK8 plays a central role.
“Through chemical proteomics and biochemical experiments, we discovered that heme interacted with DOCK8, which led to the permanent activation of its downstream target, Cdc42, with deleterious effects,” Dr Knapp said.
In the presence of heme, the cytoskeletal resilience was lost, as filaments grew rampant in all directions, resulting in macrophage paralysis. The cells lost their ability to shape-shift and could no longer chase down and engulf the invading bacteria, allowing the bacteria to multiply virtually unrestricted.
However, Dr Knapp and her colleagues found that an antimalarial drug can restore the functionality of these paralyzed macrophages.
“Quinine, which is clinically used to treat malaria and is suspected to bind heme, blocks the interaction of heme with DOCK8 and thereby improves the outcome from sepsis,” Dr Knapp said.
“This is very promising. We conclusively demonstrate that it is indeed feasible to therapeutically ‘protect’ immune cells and to restore the body’s immune defense against bacteria in hemolytic conditions.” ![]()
Docs may be uninformed about biosimilars
(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf

(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf

(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
Team develops multicolor electron microscopy
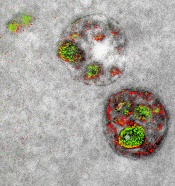
of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.” ![]()

of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.” ![]()

of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.” ![]()
Studies reveal markers of malaria resistance
Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.” ![]()

Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.” ![]()

Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.” ![]()
Study reveals ‘high-traffic’ routes of malaria importation
Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()

Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()

Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()
Tobacco plants used to manufacture malaria drug

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.” ![]()

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.” ![]()

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.”
Improving cognitive function in cancer survivors
chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”

chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”

chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”
FDA issues warning about radiation therapy devices
to receive radiation
Photo by Rhoda Baer
The US Food and Drug Administration (FDA) has warned healthcare providers to stop using radiation therapy devices manufactured and sold by Multidata Systems International Corporation.
The FDA said the company has distributed at least 2 devices in the US that were never reviewed by or registered with the FDA—(1) accessories to radiation therapy devices including the Real Time Dosimetry Waterphantom System and (2) the Dual Channel Electrometer.
Since 2003, Multidata has been under a decree that prohibits the company from designing, manufacturing, processing, and distributing medical devices, among other restrictions.
However, the FDA learned that Multidata manufactured and distributed medical devices in violation of the decree, including repairing and exchanging Waterphantom devices for newer design models.
As a result, on March 3, 2016, the FDA sent a letter to Multidata ordering the firm to stop designing, manufacturing, processing, packing, repacking, labeling, installing, holding for sale, and distributing any medical device. The company has permanently ceased operations and will be dissolving.
The FDA said it doesn’t know how many devices manufactured by Multidata are in use throughout the US or if any of these devices have caused adverse events since the decree was issued.
The agency has urged healthcare providers to stop using and dispose of any devices manufactured by Multidata.
Instead, providers should use accessories to radiation therapy devices and radiation treatment planning software that have been cleared by the FDA. Registered manufacturers of such products are listed under the IYE product code in the FDA’s Registration and Listing Database.
The FDA has also recommended that providers report any adverse events related to Multidata devices. Reports can be submitted through MedWatch, the FDA’s Safety Information and Adverse Event Reporting Program.

to receive radiation
Photo by Rhoda Baer
The US Food and Drug Administration (FDA) has warned healthcare providers to stop using radiation therapy devices manufactured and sold by Multidata Systems International Corporation.
The FDA said the company has distributed at least 2 devices in the US that were never reviewed by or registered with the FDA—(1) accessories to radiation therapy devices including the Real Time Dosimetry Waterphantom System and (2) the Dual Channel Electrometer.
Since 2003, Multidata has been under a decree that prohibits the company from designing, manufacturing, processing, and distributing medical devices, among other restrictions.
However, the FDA learned that Multidata manufactured and distributed medical devices in violation of the decree, including repairing and exchanging Waterphantom devices for newer design models.
As a result, on March 3, 2016, the FDA sent a letter to Multidata ordering the firm to stop designing, manufacturing, processing, packing, repacking, labeling, installing, holding for sale, and distributing any medical device. The company has permanently ceased operations and will be dissolving.
The FDA said it doesn’t know how many devices manufactured by Multidata are in use throughout the US or if any of these devices have caused adverse events since the decree was issued.
The agency has urged healthcare providers to stop using and dispose of any devices manufactured by Multidata.
Instead, providers should use accessories to radiation therapy devices and radiation treatment planning software that have been cleared by the FDA. Registered manufacturers of such products are listed under the IYE product code in the FDA’s Registration and Listing Database.
The FDA has also recommended that providers report any adverse events related to Multidata devices. Reports can be submitted through MedWatch, the FDA’s Safety Information and Adverse Event Reporting Program.

to receive radiation
Photo by Rhoda Baer
The US Food and Drug Administration (FDA) has warned healthcare providers to stop using radiation therapy devices manufactured and sold by Multidata Systems International Corporation.
The FDA said the company has distributed at least 2 devices in the US that were never reviewed by or registered with the FDA—(1) accessories to radiation therapy devices including the Real Time Dosimetry Waterphantom System and (2) the Dual Channel Electrometer.
Since 2003, Multidata has been under a decree that prohibits the company from designing, manufacturing, processing, and distributing medical devices, among other restrictions.
However, the FDA learned that Multidata manufactured and distributed medical devices in violation of the decree, including repairing and exchanging Waterphantom devices for newer design models.
As a result, on March 3, 2016, the FDA sent a letter to Multidata ordering the firm to stop designing, manufacturing, processing, packing, repacking, labeling, installing, holding for sale, and distributing any medical device. The company has permanently ceased operations and will be dissolving.
The FDA said it doesn’t know how many devices manufactured by Multidata are in use throughout the US or if any of these devices have caused adverse events since the decree was issued.
The agency has urged healthcare providers to stop using and dispose of any devices manufactured by Multidata.
Instead, providers should use accessories to radiation therapy devices and radiation treatment planning software that have been cleared by the FDA. Registered manufacturers of such products are listed under the IYE product code in the FDA’s Registration and Listing Database.
The FDA has also recommended that providers report any adverse events related to Multidata devices. Reports can be submitted through MedWatch, the FDA’s Safety Information and Adverse Event Reporting Program.
Burden of cancer varies by cancer type, race
Photo by Rhoda Baer
A new study suggests that leukemia and non-Hodgkin lymphoma (NHL) are among the top 10 cancers with the greatest burden (most years of healthy life lost) in the US.
The research also showed that the burden of different cancer types varied between patients belonging to different racial/ethnic groups.
For example, the contribution of leukemia to the overall cancer burden was twice as high in Hispanics as it was in non-Hispanic blacks. The same was true for NHL.
Joannie Lortet-Tieulent, of the American Cancer Society in Atlanta, Georgia, and her colleagues conducted this study and reported the results in the American Journal of Preventive Medicine.
The researchers calculated the burden of cancer in the US in 2011 for 24 cancer types. They calculated burden using disability-adjusted life years (DALYs), which combine cancer incidence, mortality, survival, and quality of life into a summary indicator.
The results suggested the burden of cancer in 2011 was over 9.8 million DALYs, which was equally shared between men and women—4.9 million DALYs for each sex.
DALYs lost to cancer were mostly related to premature death due to cancer (91%). The remaining 9% were related to impaired quality of life because of the disease or its treatment, or other disease-related issues.
Top 10 contributors
The researchers calculated the proportion of DALYs lost for each of the cancer types. And they found that lung cancer was the largest contributor to the loss of healthy years, accounting for 24% of the burden (2.4 million DALYs).
The second biggest contributor to the loss of healthy years was breast cancer (10%), followed by colorectal cancer (9%), pancreatic cancer (6%), prostate cancer (5%), leukemia (4%), liver cancer (4%), brain cancer (3%), NHL (3%), and ovarian cancer (3%).
The researchers also calculated the proportion of DALYs lost from the top 10 cancer types according to race/ethnicity.
They found the contribution of leukemia to the loss of healthy years was greatest for Hispanics (6%), followed by non-Hispanic Asians (5%), non-Hispanic whites (4%), and non-Hispanic blacks (3%).
The contribution of NHL to the loss of healthy years was greatest for Hispanics (4%), followed by non-Hispanic Asians/non-Hispanic whites (3% for both), and non-Hispanic blacks (2%).
DALYs by race/ethnicity
The researchers found that, overall, the cancer burden was highest in non-Hispanic blacks, followed by non-Hispanic whites, Hispanics, and non-Hispanic Asians. However, this pattern was not consistent across the different cancer types.
Age-standardized DALYs lost (per 100,000 individuals) were as follows:
All cancers combined
3588 for non-Hispanic blacks
2898 for non-Hispanic whites
1978 for Hispanics
1798 for non-Hispanic Asians.
Leukemia
115 for non-Hispanic blacks and non-Hispanic whites
98 for Hispanics
82 for non-Hispanic Asians.
NHL
93 for non-Hispanic whites
86 for non-Hispanic blacks
78 for Hispanics
60 for non-Hispanic Asians.
Hodgkin lymphoma
11 for non-Hispanic blacks
10 for non-Hispanic whites and Hispanics
3 for non-Hispanic Asians.
Myeloma
93 for non-Hispanic blacks
43 for non-Hispanic whites
42 for Hispanics
26 for non-Hispanic Asians.
The researchers noted that, despite these differences, the cancer burden in all races/ethnicities was driven by years of life lost. They said this highlights the need to prevent deaths by improving prevention, early detection, and treatment of cancers.

Photo by Rhoda Baer
A new study suggests that leukemia and non-Hodgkin lymphoma (NHL) are among the top 10 cancers with the greatest burden (most years of healthy life lost) in the US.
The research also showed that the burden of different cancer types varied between patients belonging to different racial/ethnic groups.
For example, the contribution of leukemia to the overall cancer burden was twice as high in Hispanics as it was in non-Hispanic blacks. The same was true for NHL.
Joannie Lortet-Tieulent, of the American Cancer Society in Atlanta, Georgia, and her colleagues conducted this study and reported the results in the American Journal of Preventive Medicine.
The researchers calculated the burden of cancer in the US in 2011 for 24 cancer types. They calculated burden using disability-adjusted life years (DALYs), which combine cancer incidence, mortality, survival, and quality of life into a summary indicator.
The results suggested the burden of cancer in 2011 was over 9.8 million DALYs, which was equally shared between men and women—4.9 million DALYs for each sex.
DALYs lost to cancer were mostly related to premature death due to cancer (91%). The remaining 9% were related to impaired quality of life because of the disease or its treatment, or other disease-related issues.
Top 10 contributors
The researchers calculated the proportion of DALYs lost for each of the cancer types. And they found that lung cancer was the largest contributor to the loss of healthy years, accounting for 24% of the burden (2.4 million DALYs).
The second biggest contributor to the loss of healthy years was breast cancer (10%), followed by colorectal cancer (9%), pancreatic cancer (6%), prostate cancer (5%), leukemia (4%), liver cancer (4%), brain cancer (3%), NHL (3%), and ovarian cancer (3%).
The researchers also calculated the proportion of DALYs lost from the top 10 cancer types according to race/ethnicity.
They found the contribution of leukemia to the loss of healthy years was greatest for Hispanics (6%), followed by non-Hispanic Asians (5%), non-Hispanic whites (4%), and non-Hispanic blacks (3%).
The contribution of NHL to the loss of healthy years was greatest for Hispanics (4%), followed by non-Hispanic Asians/non-Hispanic whites (3% for both), and non-Hispanic blacks (2%).
DALYs by race/ethnicity
The researchers found that, overall, the cancer burden was highest in non-Hispanic blacks, followed by non-Hispanic whites, Hispanics, and non-Hispanic Asians. However, this pattern was not consistent across the different cancer types.
Age-standardized DALYs lost (per 100,000 individuals) were as follows:
All cancers combined
3588 for non-Hispanic blacks
2898 for non-Hispanic whites
1978 for Hispanics
1798 for non-Hispanic Asians.
Leukemia
115 for non-Hispanic blacks and non-Hispanic whites
98 for Hispanics
82 for non-Hispanic Asians.
NHL
93 for non-Hispanic whites
86 for non-Hispanic blacks
78 for Hispanics
60 for non-Hispanic Asians.
Hodgkin lymphoma
11 for non-Hispanic blacks
10 for non-Hispanic whites and Hispanics
3 for non-Hispanic Asians.
Myeloma
93 for non-Hispanic blacks
43 for non-Hispanic whites
42 for Hispanics
26 for non-Hispanic Asians.
The researchers noted that, despite these differences, the cancer burden in all races/ethnicities was driven by years of life lost. They said this highlights the need to prevent deaths by improving prevention, early detection, and treatment of cancers.

Photo by Rhoda Baer
A new study suggests that leukemia and non-Hodgkin lymphoma (NHL) are among the top 10 cancers with the greatest burden (most years of healthy life lost) in the US.
The research also showed that the burden of different cancer types varied between patients belonging to different racial/ethnic groups.
For example, the contribution of leukemia to the overall cancer burden was twice as high in Hispanics as it was in non-Hispanic blacks. The same was true for NHL.
Joannie Lortet-Tieulent, of the American Cancer Society in Atlanta, Georgia, and her colleagues conducted this study and reported the results in the American Journal of Preventive Medicine.
The researchers calculated the burden of cancer in the US in 2011 for 24 cancer types. They calculated burden using disability-adjusted life years (DALYs), which combine cancer incidence, mortality, survival, and quality of life into a summary indicator.
The results suggested the burden of cancer in 2011 was over 9.8 million DALYs, which was equally shared between men and women—4.9 million DALYs for each sex.
DALYs lost to cancer were mostly related to premature death due to cancer (91%). The remaining 9% were related to impaired quality of life because of the disease or its treatment, or other disease-related issues.
Top 10 contributors
The researchers calculated the proportion of DALYs lost for each of the cancer types. And they found that lung cancer was the largest contributor to the loss of healthy years, accounting for 24% of the burden (2.4 million DALYs).
The second biggest contributor to the loss of healthy years was breast cancer (10%), followed by colorectal cancer (9%), pancreatic cancer (6%), prostate cancer (5%), leukemia (4%), liver cancer (4%), brain cancer (3%), NHL (3%), and ovarian cancer (3%).
The researchers also calculated the proportion of DALYs lost from the top 10 cancer types according to race/ethnicity.
They found the contribution of leukemia to the loss of healthy years was greatest for Hispanics (6%), followed by non-Hispanic Asians (5%), non-Hispanic whites (4%), and non-Hispanic blacks (3%).
The contribution of NHL to the loss of healthy years was greatest for Hispanics (4%), followed by non-Hispanic Asians/non-Hispanic whites (3% for both), and non-Hispanic blacks (2%).
DALYs by race/ethnicity
The researchers found that, overall, the cancer burden was highest in non-Hispanic blacks, followed by non-Hispanic whites, Hispanics, and non-Hispanic Asians. However, this pattern was not consistent across the different cancer types.
Age-standardized DALYs lost (per 100,000 individuals) were as follows:
All cancers combined
3588 for non-Hispanic blacks
2898 for non-Hispanic whites
1978 for Hispanics
1798 for non-Hispanic Asians.
Leukemia
115 for non-Hispanic blacks and non-Hispanic whites
98 for Hispanics
82 for non-Hispanic Asians.
NHL
93 for non-Hispanic whites
86 for non-Hispanic blacks
78 for Hispanics
60 for non-Hispanic Asians.
Hodgkin lymphoma
11 for non-Hispanic blacks
10 for non-Hispanic whites and Hispanics
3 for non-Hispanic Asians.
Myeloma
93 for non-Hispanic blacks
43 for non-Hispanic whites
42 for Hispanics
26 for non-Hispanic Asians.
The researchers noted that, despite these differences, the cancer burden in all races/ethnicities was driven by years of life lost. They said this highlights the need to prevent deaths by improving prevention, early detection, and treatment of cancers.