User login
US cancer centers spend millions in advertising
chemotherapy
Photo by Rhoda Baer
US cancer centers have substantially increased spending for consumer-directed advertising in recent years, according to researchers.
The team looked at consumer advertising for 890 cancer centers and found that total advertising spending for these centers increased from $54,229,849 in 2005 to $173,510,900 in 2014.
The researchers reported these findings in a letter to JAMA Internal Medicine.
The team analyzed data from an agency that tracks the content and number of advertisements and calculates expenditures. An advertiser was considered a cancer center if its name contained certain key words.
Advertising expenditure data covered 6 media outlets: television, magazines, radio, newspapers, billboards, and the Internet.
According to these data, from 2005 to 2014, 890 cancer centers advertised to the public. In general, inflation-adjusted spending increased for all of the types of advertising the researchers analyzed.
The team said the greatest relative growth in spending was for Internet display advertisements, which increased from less than 1% of total advertising spending in 2005 ($302,030/$54,229,849) to 5% in 2014 ($8,633,000/$173,510,900).
The researchers also found that, in 2014, 20 cancer centers accounted for 86% of the roughly $174 million total advertising spending.
Cancer Treatment Centers of America spent the most (at $101.7 million), followed by MD Anderson Cancer Center (at $13.9 million) and Memorial Sloan Kettering Cancer Center (at $9.1 million).
Among the 20 centers, 5 were for-profit, 17 were Commission on Cancer-accredited, and 9 were National Cancer Institute-designated.
The researchers said their findings likely underestimate advertising spending because the available data didn’t include all types of advertising, such as ads in cancer-specific magazines.
The team also said the effect of cancer-center advertising on the quality and costs of cancer care should be investigated. ![]()

chemotherapy
Photo by Rhoda Baer
US cancer centers have substantially increased spending for consumer-directed advertising in recent years, according to researchers.
The team looked at consumer advertising for 890 cancer centers and found that total advertising spending for these centers increased from $54,229,849 in 2005 to $173,510,900 in 2014.
The researchers reported these findings in a letter to JAMA Internal Medicine.
The team analyzed data from an agency that tracks the content and number of advertisements and calculates expenditures. An advertiser was considered a cancer center if its name contained certain key words.
Advertising expenditure data covered 6 media outlets: television, magazines, radio, newspapers, billboards, and the Internet.
According to these data, from 2005 to 2014, 890 cancer centers advertised to the public. In general, inflation-adjusted spending increased for all of the types of advertising the researchers analyzed.
The team said the greatest relative growth in spending was for Internet display advertisements, which increased from less than 1% of total advertising spending in 2005 ($302,030/$54,229,849) to 5% in 2014 ($8,633,000/$173,510,900).
The researchers also found that, in 2014, 20 cancer centers accounted for 86% of the roughly $174 million total advertising spending.
Cancer Treatment Centers of America spent the most (at $101.7 million), followed by MD Anderson Cancer Center (at $13.9 million) and Memorial Sloan Kettering Cancer Center (at $9.1 million).
Among the 20 centers, 5 were for-profit, 17 were Commission on Cancer-accredited, and 9 were National Cancer Institute-designated.
The researchers said their findings likely underestimate advertising spending because the available data didn’t include all types of advertising, such as ads in cancer-specific magazines.
The team also said the effect of cancer-center advertising on the quality and costs of cancer care should be investigated. ![]()

chemotherapy
Photo by Rhoda Baer
US cancer centers have substantially increased spending for consumer-directed advertising in recent years, according to researchers.
The team looked at consumer advertising for 890 cancer centers and found that total advertising spending for these centers increased from $54,229,849 in 2005 to $173,510,900 in 2014.
The researchers reported these findings in a letter to JAMA Internal Medicine.
The team analyzed data from an agency that tracks the content and number of advertisements and calculates expenditures. An advertiser was considered a cancer center if its name contained certain key words.
Advertising expenditure data covered 6 media outlets: television, magazines, radio, newspapers, billboards, and the Internet.
According to these data, from 2005 to 2014, 890 cancer centers advertised to the public. In general, inflation-adjusted spending increased for all of the types of advertising the researchers analyzed.
The team said the greatest relative growth in spending was for Internet display advertisements, which increased from less than 1% of total advertising spending in 2005 ($302,030/$54,229,849) to 5% in 2014 ($8,633,000/$173,510,900).
The researchers also found that, in 2014, 20 cancer centers accounted for 86% of the roughly $174 million total advertising spending.
Cancer Treatment Centers of America spent the most (at $101.7 million), followed by MD Anderson Cancer Center (at $13.9 million) and Memorial Sloan Kettering Cancer Center (at $9.1 million).
Among the 20 centers, 5 were for-profit, 17 were Commission on Cancer-accredited, and 9 were National Cancer Institute-designated.
The researchers said their findings likely underestimate advertising spending because the available data didn’t include all types of advertising, such as ads in cancer-specific magazines.
The team also said the effect of cancer-center advertising on the quality and costs of cancer care should be investigated. ![]()
Study reveals global variations of P vivax
from patients in Thailand
Image by Wanlapa Roobsoong
Researchers say they have uncovered the global, evolving, and historic make-up of the malaria parasite Plasmodium vivax.
The group’s study revealed 4 genetically distinct populations of P vivax that provide insight into the movement of the parasite over time and
show how it is still adapting to regional variations in the mosquitoes that transmit P vivax, the humans infected with the parasite, and the drugs used to fight it.
“Our findings show it is evolving in response to antimalarial drugs and adapting to regional differences, indicating a wide range of approaches will likely be necessary to eliminate it globally,” said Jane Carlton, PhD, of New York University in New York, New York.
Dr Carlton and her colleagues reported these findings in Nature Genetics.
The researchers sequenced 182 DNA samples of P vivax collected from patients in 11 countries. The team said this provided new insights into the nature of P vivax as it exists today and also served as a “genetic history book” of the studied regions.
“The DNA data show that P vivax has clearly had a different history of association with global human populations than other malaria parasites, indicating that unique aspects of its biology may have influenced the ways in which it spread around the world,” said Daniel Neafsey, PhD, of the Broad Institute in Cambridge, Massachusetts.
Specifically, the researchers found that Central and South American P vivax populations are genetically diverse and distinct from all other contemporary P vivax populations. The team said this suggests that New World parasites may have been introduced by colonial seafarers and represent a now-eliminated European parasite population.
The researchers also found that contemporary African and South Asian P vivax populations are genetically similar. They said this suggests that South Asian P vivax populations may have genetically mingled with European lineages during the colonial era, or it may reflect ancient connections between human populations in the Eastern Mediterranean, Middle East, and Indian subcontinent.
Another finding was the relatively homogeneous genetic makeup of P vivax in Mexico, which reflects a steady decline of the disease in this country over the last decade.
By contrast, the Papua New Guinea population of P vivax was shown to be very diverse relative to other P vivax populations.
A similar study, which also illustrated the global variations of P vivax, was recently published in Nature Genetics in as well. ![]()

from patients in Thailand
Image by Wanlapa Roobsoong
Researchers say they have uncovered the global, evolving, and historic make-up of the malaria parasite Plasmodium vivax.
The group’s study revealed 4 genetically distinct populations of P vivax that provide insight into the movement of the parasite over time and
show how it is still adapting to regional variations in the mosquitoes that transmit P vivax, the humans infected with the parasite, and the drugs used to fight it.
“Our findings show it is evolving in response to antimalarial drugs and adapting to regional differences, indicating a wide range of approaches will likely be necessary to eliminate it globally,” said Jane Carlton, PhD, of New York University in New York, New York.
Dr Carlton and her colleagues reported these findings in Nature Genetics.
The researchers sequenced 182 DNA samples of P vivax collected from patients in 11 countries. The team said this provided new insights into the nature of P vivax as it exists today and also served as a “genetic history book” of the studied regions.
“The DNA data show that P vivax has clearly had a different history of association with global human populations than other malaria parasites, indicating that unique aspects of its biology may have influenced the ways in which it spread around the world,” said Daniel Neafsey, PhD, of the Broad Institute in Cambridge, Massachusetts.
Specifically, the researchers found that Central and South American P vivax populations are genetically diverse and distinct from all other contemporary P vivax populations. The team said this suggests that New World parasites may have been introduced by colonial seafarers and represent a now-eliminated European parasite population.
The researchers also found that contemporary African and South Asian P vivax populations are genetically similar. They said this suggests that South Asian P vivax populations may have genetically mingled with European lineages during the colonial era, or it may reflect ancient connections between human populations in the Eastern Mediterranean, Middle East, and Indian subcontinent.
Another finding was the relatively homogeneous genetic makeup of P vivax in Mexico, which reflects a steady decline of the disease in this country over the last decade.
By contrast, the Papua New Guinea population of P vivax was shown to be very diverse relative to other P vivax populations.
A similar study, which also illustrated the global variations of P vivax, was recently published in Nature Genetics in as well. ![]()

from patients in Thailand
Image by Wanlapa Roobsoong
Researchers say they have uncovered the global, evolving, and historic make-up of the malaria parasite Plasmodium vivax.
The group’s study revealed 4 genetically distinct populations of P vivax that provide insight into the movement of the parasite over time and
show how it is still adapting to regional variations in the mosquitoes that transmit P vivax, the humans infected with the parasite, and the drugs used to fight it.
“Our findings show it is evolving in response to antimalarial drugs and adapting to regional differences, indicating a wide range of approaches will likely be necessary to eliminate it globally,” said Jane Carlton, PhD, of New York University in New York, New York.
Dr Carlton and her colleagues reported these findings in Nature Genetics.
The researchers sequenced 182 DNA samples of P vivax collected from patients in 11 countries. The team said this provided new insights into the nature of P vivax as it exists today and also served as a “genetic history book” of the studied regions.
“The DNA data show that P vivax has clearly had a different history of association with global human populations than other malaria parasites, indicating that unique aspects of its biology may have influenced the ways in which it spread around the world,” said Daniel Neafsey, PhD, of the Broad Institute in Cambridge, Massachusetts.
Specifically, the researchers found that Central and South American P vivax populations are genetically diverse and distinct from all other contemporary P vivax populations. The team said this suggests that New World parasites may have been introduced by colonial seafarers and represent a now-eliminated European parasite population.
The researchers also found that contemporary African and South Asian P vivax populations are genetically similar. They said this suggests that South Asian P vivax populations may have genetically mingled with European lineages during the colonial era, or it may reflect ancient connections between human populations in the Eastern Mediterranean, Middle East, and Indian subcontinent.
Another finding was the relatively homogeneous genetic makeup of P vivax in Mexico, which reflects a steady decline of the disease in this country over the last decade.
By contrast, the Papua New Guinea population of P vivax was shown to be very diverse relative to other P vivax populations.
A similar study, which also illustrated the global variations of P vivax, was recently published in Nature Genetics in as well. ![]()
How multiple infections make malaria worse
Image by Ute Frevert
and Margaret Shear
New research suggests that infections with 2 types of malaria parasite lead to greater health risks because 1 species helps the other thrive.
Investigators sought to understand what happens when the 2 most common malaria parasites cause infection at the same time, as they are known to attack the body in different ways.
The team found the first parasite helps provide the second with more of the resources it needs to prosper.
“Immune responses are assumed to determine the outcome of interactions between parasite species, but our study clearly shows that resources can be more important,” said Sarah Reece, of the University of Edinburgh in Scotland.
“Our findings also challenge ideas that 1 species will outcompete the other, which explains why infections involving 2 parasite species can pose a greater health risk to patients.”
Dr Reece and her colleagues recounted these findings in Ecology Letters.
In humans, the malaria parasite Plasmodium falciparum infects red blood cells of all ages, while the Plasmodium vivax parasite attacks only young red blood cells.
The current study, conducted in mice with equivalent malaria parasites (P chabaudi and P yoelii), showed that the body’s response to the first infection produces more of the type of red blood cell the second parasite needs.
In response to the first infection, millions of red blood cells are destroyed. The body responds by replenishing these cells.
The fresh cells then become infected by the second type of parasite, making the infection worse.
The investigators said these results appear to explain why infections with both P falciparum and P vivax often have worse outcomes for patients than infections with a single malaria parasite. ![]()

Image by Ute Frevert
and Margaret Shear
New research suggests that infections with 2 types of malaria parasite lead to greater health risks because 1 species helps the other thrive.
Investigators sought to understand what happens when the 2 most common malaria parasites cause infection at the same time, as they are known to attack the body in different ways.
The team found the first parasite helps provide the second with more of the resources it needs to prosper.
“Immune responses are assumed to determine the outcome of interactions between parasite species, but our study clearly shows that resources can be more important,” said Sarah Reece, of the University of Edinburgh in Scotland.
“Our findings also challenge ideas that 1 species will outcompete the other, which explains why infections involving 2 parasite species can pose a greater health risk to patients.”
Dr Reece and her colleagues recounted these findings in Ecology Letters.
In humans, the malaria parasite Plasmodium falciparum infects red blood cells of all ages, while the Plasmodium vivax parasite attacks only young red blood cells.
The current study, conducted in mice with equivalent malaria parasites (P chabaudi and P yoelii), showed that the body’s response to the first infection produces more of the type of red blood cell the second parasite needs.
In response to the first infection, millions of red blood cells are destroyed. The body responds by replenishing these cells.
The fresh cells then become infected by the second type of parasite, making the infection worse.
The investigators said these results appear to explain why infections with both P falciparum and P vivax often have worse outcomes for patients than infections with a single malaria parasite. ![]()

Image by Ute Frevert
and Margaret Shear
New research suggests that infections with 2 types of malaria parasite lead to greater health risks because 1 species helps the other thrive.
Investigators sought to understand what happens when the 2 most common malaria parasites cause infection at the same time, as they are known to attack the body in different ways.
The team found the first parasite helps provide the second with more of the resources it needs to prosper.
“Immune responses are assumed to determine the outcome of interactions between parasite species, but our study clearly shows that resources can be more important,” said Sarah Reece, of the University of Edinburgh in Scotland.
“Our findings also challenge ideas that 1 species will outcompete the other, which explains why infections involving 2 parasite species can pose a greater health risk to patients.”
Dr Reece and her colleagues recounted these findings in Ecology Letters.
In humans, the malaria parasite Plasmodium falciparum infects red blood cells of all ages, while the Plasmodium vivax parasite attacks only young red blood cells.
The current study, conducted in mice with equivalent malaria parasites (P chabaudi and P yoelii), showed that the body’s response to the first infection produces more of the type of red blood cell the second parasite needs.
In response to the first infection, millions of red blood cells are destroyed. The body responds by replenishing these cells.
The fresh cells then become infected by the second type of parasite, making the infection worse.
The investigators said these results appear to explain why infections with both P falciparum and P vivax often have worse outcomes for patients than infections with a single malaria parasite. ![]()
Efficacy of malaria vaccine declines over time
Photo by Caitlin Kleiboer
Results from a phase 2 study of the malaria vaccine RTS,S (also known as RTS,S/AS01 or Mosquirix) suggest its efficacy decreases over time, and this decline is fastest in children living in areas with higher-than-average rates of malaria.
Researchers say the results suggest the benefits of the vaccine are likely to vary across different populations and highlight the need for more research to
determine the most effective way of using RTS,S, which last year became the first malaria vaccine to receive a green light from the European Medicines Agency.
“We found that 3-dose vaccination with RTS,S was initially protective, but this was offset by a rebound in later years among children exposed to higher-than-average levels of malaria-carrying mosquitoes,” said Philip Bejon, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme in Kilifi, Kenya.
Dr Bejon and his colleagues reported these results in NEJM.
The researchers followed 447 children who had received 3 doses of either RTS,S or a rabies (control) vaccine when they were 5 months to 17 months old.
After 7 years, there were 312 children still involved in the study. During the first year, the risk of getting malaria in the vaccinated children was 35.9% less than in the control group. After 7 years, this protection fell to 3.6%.
And in children exposed to higher-than-average rates of malaria, there were slightly more cases of malaria in the vaccinated group than the control group—1002 and 992 cases, respectively—5 years after vaccination.
This “rebound” effect, which has been seen in previous studies, is thought to occur because children initially protected by the vaccine develop their natural immunity against malaria more slowly than unvaccinated children.
Results from a phase 3 study showed that 3 doses of RTS,S reduced the risk of malaria in young children by 28% over 4 years, but this improved to 36% when children were given a fourth dose 18 months after the first dose. Longer-term follow up of these children is ongoing.
“Overall, our study shows that RTS,S can benefit children but suggests that a fourth dose may be important for sustaining this protection over the long term and to protect against a potential rebound,” said Ally Olotu, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme.
“Results from 3 sites involved in the original phase 3 study that are continuing follow up, and the WHO’s planned pilot program, will tell us more about the vaccine’s efficacy in different settings and help determine which populations would benefit most from receiving it as part of a wider vaccination strategy.” ![]()

Photo by Caitlin Kleiboer
Results from a phase 2 study of the malaria vaccine RTS,S (also known as RTS,S/AS01 or Mosquirix) suggest its efficacy decreases over time, and this decline is fastest in children living in areas with higher-than-average rates of malaria.
Researchers say the results suggest the benefits of the vaccine are likely to vary across different populations and highlight the need for more research to
determine the most effective way of using RTS,S, which last year became the first malaria vaccine to receive a green light from the European Medicines Agency.
“We found that 3-dose vaccination with RTS,S was initially protective, but this was offset by a rebound in later years among children exposed to higher-than-average levels of malaria-carrying mosquitoes,” said Philip Bejon, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme in Kilifi, Kenya.
Dr Bejon and his colleagues reported these results in NEJM.
The researchers followed 447 children who had received 3 doses of either RTS,S or a rabies (control) vaccine when they were 5 months to 17 months old.
After 7 years, there were 312 children still involved in the study. During the first year, the risk of getting malaria in the vaccinated children was 35.9% less than in the control group. After 7 years, this protection fell to 3.6%.
And in children exposed to higher-than-average rates of malaria, there were slightly more cases of malaria in the vaccinated group than the control group—1002 and 992 cases, respectively—5 years after vaccination.
This “rebound” effect, which has been seen in previous studies, is thought to occur because children initially protected by the vaccine develop their natural immunity against malaria more slowly than unvaccinated children.
Results from a phase 3 study showed that 3 doses of RTS,S reduced the risk of malaria in young children by 28% over 4 years, but this improved to 36% when children were given a fourth dose 18 months after the first dose. Longer-term follow up of these children is ongoing.
“Overall, our study shows that RTS,S can benefit children but suggests that a fourth dose may be important for sustaining this protection over the long term and to protect against a potential rebound,” said Ally Olotu, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme.
“Results from 3 sites involved in the original phase 3 study that are continuing follow up, and the WHO’s planned pilot program, will tell us more about the vaccine’s efficacy in different settings and help determine which populations would benefit most from receiving it as part of a wider vaccination strategy.” ![]()

Photo by Caitlin Kleiboer
Results from a phase 2 study of the malaria vaccine RTS,S (also known as RTS,S/AS01 or Mosquirix) suggest its efficacy decreases over time, and this decline is fastest in children living in areas with higher-than-average rates of malaria.
Researchers say the results suggest the benefits of the vaccine are likely to vary across different populations and highlight the need for more research to
determine the most effective way of using RTS,S, which last year became the first malaria vaccine to receive a green light from the European Medicines Agency.
“We found that 3-dose vaccination with RTS,S was initially protective, but this was offset by a rebound in later years among children exposed to higher-than-average levels of malaria-carrying mosquitoes,” said Philip Bejon, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme in Kilifi, Kenya.
Dr Bejon and his colleagues reported these results in NEJM.
The researchers followed 447 children who had received 3 doses of either RTS,S or a rabies (control) vaccine when they were 5 months to 17 months old.
After 7 years, there were 312 children still involved in the study. During the first year, the risk of getting malaria in the vaccinated children was 35.9% less than in the control group. After 7 years, this protection fell to 3.6%.
And in children exposed to higher-than-average rates of malaria, there were slightly more cases of malaria in the vaccinated group than the control group—1002 and 992 cases, respectively—5 years after vaccination.
This “rebound” effect, which has been seen in previous studies, is thought to occur because children initially protected by the vaccine develop their natural immunity against malaria more slowly than unvaccinated children.
Results from a phase 3 study showed that 3 doses of RTS,S reduced the risk of malaria in young children by 28% over 4 years, but this improved to 36% when children were given a fourth dose 18 months after the first dose. Longer-term follow up of these children is ongoing.
“Overall, our study shows that RTS,S can benefit children but suggests that a fourth dose may be important for sustaining this protection over the long term and to protect against a potential rebound,” said Ally Olotu, PhD, of the Kenya Medical Research Institute–Wellcome Trust Programme.
“Results from 3 sites involved in the original phase 3 study that are continuing follow up, and the WHO’s planned pilot program, will tell us more about the vaccine’s efficacy in different settings and help determine which populations would benefit most from receiving it as part of a wider vaccination strategy.” ![]()
Computer model shows how spleen filters blood
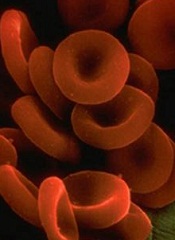
Researchers have created a computer model that shows how tiny slits in the spleen prevent old, diseased, or misshapen red blood cells from re-entering the bloodstream.
The team says the model can be used to study the spleen’s role in controlling diseases that affect the size and structure of red blood cells—such as sickle cell anemia, thalassemia, and malaria—and to develop new diagnostics and therapeutics for these diseases.
The researchers described the model in PNAS.
Previous studies have shown that part of the spleen’s filtration process relies on having red blood cells squeeze through tiny slits between the endothelial cells that line the spleen’s blood vessels. These “interendothelial slits” are no larger than 1.2 µm tall, 4 µm wide, and 1.9 µm deep.
More rigid and misshapen blood cells might not be able to pass through these narrow passages. And this process cannot be observed in vivo because of the minute size of the slits.
In order to “see” how the interendothelial slits regulate red blood cell circulation, the researchers created a computer simulation based on dissipative particle dynamics, a modeling method.
Their model allowed the team to determine the range of cell sizes and shapes that could fit through the slits. The range closely mirrored the range of sizes and shapes for healthy red blood cells, indicating that only healthy cells should be able to pass through the slits.
“The computational and analytical models from this work, along with a variety of experimental observations, point to a more detailed picture of how the physiology of the human spleen likely influences several key geometrical characteristics of red blood cells,” said study author Subra Suresh, ScD, of Carnegie Mellon University in Pittsburgh, Pennsylvania.
“They also offer better understanding of how the circulatory bottleneck for the red blood cell in the spleen could affect a variety of acute and chronic disease states arising from hereditary disorders, human cancers, and infectious diseases, with implications for therapeutic interventions and drug efficacy assays.”
In addition to giving researchers a better picture of how the spleen functions, the findings provide new insights into drug treatments.
A class of drugs currently in development for treating malaria alters the shape of red blood cells infected with malaria, theoretically preventing them from passing through interendothelial slits. One such drug, spiroindoline KAE609, is in clinical trials.
The researchers’ results might also explain why artemisinin-based antimalarial drugs, which stiffen healthy and malaria-infected red blood cells, can lead to severe anemia. ![]()

Researchers have created a computer model that shows how tiny slits in the spleen prevent old, diseased, or misshapen red blood cells from re-entering the bloodstream.
The team says the model can be used to study the spleen’s role in controlling diseases that affect the size and structure of red blood cells—such as sickle cell anemia, thalassemia, and malaria—and to develop new diagnostics and therapeutics for these diseases.
The researchers described the model in PNAS.
Previous studies have shown that part of the spleen’s filtration process relies on having red blood cells squeeze through tiny slits between the endothelial cells that line the spleen’s blood vessels. These “interendothelial slits” are no larger than 1.2 µm tall, 4 µm wide, and 1.9 µm deep.
More rigid and misshapen blood cells might not be able to pass through these narrow passages. And this process cannot be observed in vivo because of the minute size of the slits.
In order to “see” how the interendothelial slits regulate red blood cell circulation, the researchers created a computer simulation based on dissipative particle dynamics, a modeling method.
Their model allowed the team to determine the range of cell sizes and shapes that could fit through the slits. The range closely mirrored the range of sizes and shapes for healthy red blood cells, indicating that only healthy cells should be able to pass through the slits.
“The computational and analytical models from this work, along with a variety of experimental observations, point to a more detailed picture of how the physiology of the human spleen likely influences several key geometrical characteristics of red blood cells,” said study author Subra Suresh, ScD, of Carnegie Mellon University in Pittsburgh, Pennsylvania.
“They also offer better understanding of how the circulatory bottleneck for the red blood cell in the spleen could affect a variety of acute and chronic disease states arising from hereditary disorders, human cancers, and infectious diseases, with implications for therapeutic interventions and drug efficacy assays.”
In addition to giving researchers a better picture of how the spleen functions, the findings provide new insights into drug treatments.
A class of drugs currently in development for treating malaria alters the shape of red blood cells infected with malaria, theoretically preventing them from passing through interendothelial slits. One such drug, spiroindoline KAE609, is in clinical trials.
The researchers’ results might also explain why artemisinin-based antimalarial drugs, which stiffen healthy and malaria-infected red blood cells, can lead to severe anemia. ![]()

Researchers have created a computer model that shows how tiny slits in the spleen prevent old, diseased, or misshapen red blood cells from re-entering the bloodstream.
The team says the model can be used to study the spleen’s role in controlling diseases that affect the size and structure of red blood cells—such as sickle cell anemia, thalassemia, and malaria—and to develop new diagnostics and therapeutics for these diseases.
The researchers described the model in PNAS.
Previous studies have shown that part of the spleen’s filtration process relies on having red blood cells squeeze through tiny slits between the endothelial cells that line the spleen’s blood vessels. These “interendothelial slits” are no larger than 1.2 µm tall, 4 µm wide, and 1.9 µm deep.
More rigid and misshapen blood cells might not be able to pass through these narrow passages. And this process cannot be observed in vivo because of the minute size of the slits.
In order to “see” how the interendothelial slits regulate red blood cell circulation, the researchers created a computer simulation based on dissipative particle dynamics, a modeling method.
Their model allowed the team to determine the range of cell sizes and shapes that could fit through the slits. The range closely mirrored the range of sizes and shapes for healthy red blood cells, indicating that only healthy cells should be able to pass through the slits.
“The computational and analytical models from this work, along with a variety of experimental observations, point to a more detailed picture of how the physiology of the human spleen likely influences several key geometrical characteristics of red blood cells,” said study author Subra Suresh, ScD, of Carnegie Mellon University in Pittsburgh, Pennsylvania.
“They also offer better understanding of how the circulatory bottleneck for the red blood cell in the spleen could affect a variety of acute and chronic disease states arising from hereditary disorders, human cancers, and infectious diseases, with implications for therapeutic interventions and drug efficacy assays.”
In addition to giving researchers a better picture of how the spleen functions, the findings provide new insights into drug treatments.
A class of drugs currently in development for treating malaria alters the shape of red blood cells infected with malaria, theoretically preventing them from passing through interendothelial slits. One such drug, spiroindoline KAE609, is in clinical trials.
The researchers’ results might also explain why artemisinin-based antimalarial drugs, which stiffen healthy and malaria-infected red blood cells, can lead to severe anemia. ![]()
Artemisinin resistance confined to Asia, study shows
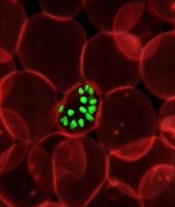
infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
The first global mapping of artemisinin resistance indicates that resistance to the drug, which is used to treat Plasmodium falciparum malaria, is confined to Southeast Asia and has not yet spread to sub-Saharan Africa.
Results of the effort, known as the KARMA study, were published in NEJM.
The study builds on the 2014 discovery that the K13 gene is the major determinant of P falciparum’s resistance to artemisinin.
Researchers studied the diversity of the K13 gene in 14,037 blood samples taken from P falciparum-infected patients in 59 malaria-endemic countries—72% in Africa, 19% in Asia, 8% in Latin America, and 1% in Oceania. All samples were collected after 2012.
The researchers identified 108 nonsynonymous K13 mutations. In Asia, 36.5% of the mutations were distributed within 2 areas—Cambodia-Vietnam-Laos and western Thailand-Myanmar-China—with no overlap.
In samples from Africa, the researchers identified nonsynonymous K13 mutations that were not associated with artemisinin resistance, including the most frequent mutation found in Africa, A578S.
“We suspect that only a small number of mutations appear to be associated with resistance, which should facilitate global monitoring of resistance to artemisinin,” said study author Odile Mercereau-Puijalon, PhD, of the Institut Pasteur in Paris, France.
“Until now, scientists have not had the tools to be properly informed about the nature of resistance to antimalarial drugs in key affected regions such as sub-Saharan Africa,” added Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia.
“We now have the capacity, thanks to molecular markers, to be able to trace—at a global level and virtually in real-time—resistance to antimalarial drugs. We must ensure that we use this technology to keep us a step ahead of the parasite.” ![]()

infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
The first global mapping of artemisinin resistance indicates that resistance to the drug, which is used to treat Plasmodium falciparum malaria, is confined to Southeast Asia and has not yet spread to sub-Saharan Africa.
Results of the effort, known as the KARMA study, were published in NEJM.
The study builds on the 2014 discovery that the K13 gene is the major determinant of P falciparum’s resistance to artemisinin.
Researchers studied the diversity of the K13 gene in 14,037 blood samples taken from P falciparum-infected patients in 59 malaria-endemic countries—72% in Africa, 19% in Asia, 8% in Latin America, and 1% in Oceania. All samples were collected after 2012.
The researchers identified 108 nonsynonymous K13 mutations. In Asia, 36.5% of the mutations were distributed within 2 areas—Cambodia-Vietnam-Laos and western Thailand-Myanmar-China—with no overlap.
In samples from Africa, the researchers identified nonsynonymous K13 mutations that were not associated with artemisinin resistance, including the most frequent mutation found in Africa, A578S.
“We suspect that only a small number of mutations appear to be associated with resistance, which should facilitate global monitoring of resistance to artemisinin,” said study author Odile Mercereau-Puijalon, PhD, of the Institut Pasteur in Paris, France.
“Until now, scientists have not had the tools to be properly informed about the nature of resistance to antimalarial drugs in key affected regions such as sub-Saharan Africa,” added Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia.
“We now have the capacity, thanks to molecular markers, to be able to trace—at a global level and virtually in real-time—resistance to antimalarial drugs. We must ensure that we use this technology to keep us a step ahead of the parasite.” ![]()

infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
The first global mapping of artemisinin resistance indicates that resistance to the drug, which is used to treat Plasmodium falciparum malaria, is confined to Southeast Asia and has not yet spread to sub-Saharan Africa.
Results of the effort, known as the KARMA study, were published in NEJM.
The study builds on the 2014 discovery that the K13 gene is the major determinant of P falciparum’s resistance to artemisinin.
Researchers studied the diversity of the K13 gene in 14,037 blood samples taken from P falciparum-infected patients in 59 malaria-endemic countries—72% in Africa, 19% in Asia, 8% in Latin America, and 1% in Oceania. All samples were collected after 2012.
The researchers identified 108 nonsynonymous K13 mutations. In Asia, 36.5% of the mutations were distributed within 2 areas—Cambodia-Vietnam-Laos and western Thailand-Myanmar-China—with no overlap.
In samples from Africa, the researchers identified nonsynonymous K13 mutations that were not associated with artemisinin resistance, including the most frequent mutation found in Africa, A578S.
“We suspect that only a small number of mutations appear to be associated with resistance, which should facilitate global monitoring of resistance to artemisinin,” said study author Odile Mercereau-Puijalon, PhD, of the Institut Pasteur in Paris, France.
“Until now, scientists have not had the tools to be properly informed about the nature of resistance to antimalarial drugs in key affected regions such as sub-Saharan Africa,” added Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia.
“We now have the capacity, thanks to molecular markers, to be able to trace—at a global level and virtually in real-time—resistance to antimalarial drugs. We must ensure that we use this technology to keep us a step ahead of the parasite.” ![]()
Agreements may constrain publication of trial results

for a clinical trial
Photo by Esther Dyson
Publication agreements between industry and academic investigators involved in clinical trials are not often reported in the publications themselves, according to a study published in PLOS Medicine.
In most of the agreements studied, industry had the right to reject or review manuscripts before publication.
Therefore, according to researchers, publication agreements may compromise the scientific evidence base established by randomized clinical trials.
Matthias Briel, MD, of the University Hospital Basel in Switzerland, and his colleagues sought to understand how publication agreements might constrain the publication of trial results.
The researchers examined publication agreements in 647 randomized trial protocols approved from 2000 to 2003 by 6 research ethics committees in Switzerland, Canada, and Germany, as well as the 388 corresponding journal publications.
The team found that 71% of protocols mentioned an agreement on publication rights between industry and academic investigators.
In 86% of those agreements, industry retained the right to disapprove or at least review manuscripts before publication.
And 74% of the agreements documented in protocols were not mentioned in corresponding journal articles.
The researchers noted that half of the included journal articles were published before 2008, leaving open the possibility that these findings do not reflect current practice.
Nonetheless, the team said the findings suggest that more transparency on publication constraints is warranted. ![]()

for a clinical trial
Photo by Esther Dyson
Publication agreements between industry and academic investigators involved in clinical trials are not often reported in the publications themselves, according to a study published in PLOS Medicine.
In most of the agreements studied, industry had the right to reject or review manuscripts before publication.
Therefore, according to researchers, publication agreements may compromise the scientific evidence base established by randomized clinical trials.
Matthias Briel, MD, of the University Hospital Basel in Switzerland, and his colleagues sought to understand how publication agreements might constrain the publication of trial results.
The researchers examined publication agreements in 647 randomized trial protocols approved from 2000 to 2003 by 6 research ethics committees in Switzerland, Canada, and Germany, as well as the 388 corresponding journal publications.
The team found that 71% of protocols mentioned an agreement on publication rights between industry and academic investigators.
In 86% of those agreements, industry retained the right to disapprove or at least review manuscripts before publication.
And 74% of the agreements documented in protocols were not mentioned in corresponding journal articles.
The researchers noted that half of the included journal articles were published before 2008, leaving open the possibility that these findings do not reflect current practice.
Nonetheless, the team said the findings suggest that more transparency on publication constraints is warranted. ![]()

for a clinical trial
Photo by Esther Dyson
Publication agreements between industry and academic investigators involved in clinical trials are not often reported in the publications themselves, according to a study published in PLOS Medicine.
In most of the agreements studied, industry had the right to reject or review manuscripts before publication.
Therefore, according to researchers, publication agreements may compromise the scientific evidence base established by randomized clinical trials.
Matthias Briel, MD, of the University Hospital Basel in Switzerland, and his colleagues sought to understand how publication agreements might constrain the publication of trial results.
The researchers examined publication agreements in 647 randomized trial protocols approved from 2000 to 2003 by 6 research ethics committees in Switzerland, Canada, and Germany, as well as the 388 corresponding journal publications.
The team found that 71% of protocols mentioned an agreement on publication rights between industry and academic investigators.
In 86% of those agreements, industry retained the right to disapprove or at least review manuscripts before publication.
And 74% of the agreements documented in protocols were not mentioned in corresponding journal articles.
The researchers noted that half of the included journal articles were published before 2008, leaving open the possibility that these findings do not reflect current practice.
Nonetheless, the team said the findings suggest that more transparency on publication constraints is warranted.
P vivax evolving differently in different regions
Plasmodium vivax
Image by Mae Melvin
Genomic research suggests the malaria parasite Plasmodium vivax is evolving rapidly to adapt to conditions in different geographic locations.
Researchers studied more than 200 parasite samples from across the Asia-Pacific region and found that P vivax has evolved differently in different areas.
The team identified substantial differences in the frequency of copy number variations (CNVs) in samples from western Thailand, western Cambodia, and Papua Indonesia.
They believe this is a result of the different antimalarial drugs used in these regions.
The researchers described this work in Nature Genetics.
“For so long, it’s not been possible to study P vivax genomes in detail, on a large-scale, but now we can, and we’re seeing the effect that drug use has on how parasites are evolving,” said study author Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in the UK.
He and his colleagues studied the genomes of 228 parasite samples, identifying the strains carried by each patient and revealing their infection history. Most samples came from Southeast Asia (Thailand, Cambodia, Vietnam, Laos, Myanmar, and Malaysia) and Oceania (Papua Indonesia and Papua New Guinea), but the team also studied samples from China, India, Sri Lanka, Brazil, and Madagascar.
The researchers performed detailed population genetic analyses using 148 samples from western Thailand, western Cambodia, and Papua Indonesia. This revealed CNVs in 9 regions of the core genome, and the frequency of the 4 most common CNVs varied greatly according to geographical location.
The first common CNV was a 9-kb deletion on chromosome 8 that includes the first 3 exons of a gene encoding a cytoadherence-linked asexual protein. The CNV was present in 73% of Papua Indonesia samples, 6% of western Cambodia samples, and 3% of western Thailand samples.
The second common CNV was a 7-kb duplication on chromosome 6 that encompasses pvdbp, the gene that encodes the Duffy-binding protein, which mediates P vivax’s invasion of erythrocytes. It was present in 5% of Papua Indonesia samples, 35% of western Cambodia samples, and 25% of western Thailand samples.
The third common CNV was a 37-kb duplication on chromosome 10 that includes pvmdr1, which has been associated with resistance to mefloquine and is homologous to the pfmdr1 amplification responsible for mefloquine resistance in P falciparum. This CNV was only present in samples from western Thailand.
The fourth common CNV was a 3-kb duplication on chromosome 14 that includes the gene PVX_101445. It was found only in Papua Indonesia samples.
“Our study shows that the strongest evidence of evolution is in Papua, Indonesia, where resistance of P vivax to chloroquine is now rampant,” said Ric Price, MD, of the University of Oxford in the UK.
“These data provide crucial information from which we can start to identify the mechanisms of drug resistance in P vivax.”
“We can see in the genome that drug resistance is a huge driver for evolution,” added Richard Pearson, PhD, of the Wellcome Trust Sanger Institute.
“Intriguingly, in some places, this process appears to be happening in response to drugs used primarily to treat a different malaria parasite, P falciparum. Although the exact cause isn’t known, this is a worrying sign that drug resistance is becoming deeply entrenched in the parasite population.”
The researchers said there are a few possible reasons why P vivax may be evolving to evade drugs used against P falciparum.
Many people carry mixed infections of both species of parasite, so, in treating one species, the other automatically gets exposed to the drug. Another culprit may be unsupervised drug use—where many people take the most readily available, rather than the most suitable, antimalarial drug.
Another finding from this study was that, when the researchers identified patients who were carrying multiple strains of parasite, the genomic data made it possible to determine how closely the different strains were related to one another.
“This means that we can now start to pull apart the genetic complexity of individual Plasmodium vivax infections and work out whether the parasites came from one or more mosquito bites,” Kwiatkowski said. “It provides a way of addressing fundamental questions about how P vivax is transmitted and how it persists within a community and, in particular, about the biology of relapsing infections.”

Plasmodium vivax
Image by Mae Melvin
Genomic research suggests the malaria parasite Plasmodium vivax is evolving rapidly to adapt to conditions in different geographic locations.
Researchers studied more than 200 parasite samples from across the Asia-Pacific region and found that P vivax has evolved differently in different areas.
The team identified substantial differences in the frequency of copy number variations (CNVs) in samples from western Thailand, western Cambodia, and Papua Indonesia.
They believe this is a result of the different antimalarial drugs used in these regions.
The researchers described this work in Nature Genetics.
“For so long, it’s not been possible to study P vivax genomes in detail, on a large-scale, but now we can, and we’re seeing the effect that drug use has on how parasites are evolving,” said study author Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in the UK.
He and his colleagues studied the genomes of 228 parasite samples, identifying the strains carried by each patient and revealing their infection history. Most samples came from Southeast Asia (Thailand, Cambodia, Vietnam, Laos, Myanmar, and Malaysia) and Oceania (Papua Indonesia and Papua New Guinea), but the team also studied samples from China, India, Sri Lanka, Brazil, and Madagascar.
The researchers performed detailed population genetic analyses using 148 samples from western Thailand, western Cambodia, and Papua Indonesia. This revealed CNVs in 9 regions of the core genome, and the frequency of the 4 most common CNVs varied greatly according to geographical location.
The first common CNV was a 9-kb deletion on chromosome 8 that includes the first 3 exons of a gene encoding a cytoadherence-linked asexual protein. The CNV was present in 73% of Papua Indonesia samples, 6% of western Cambodia samples, and 3% of western Thailand samples.
The second common CNV was a 7-kb duplication on chromosome 6 that encompasses pvdbp, the gene that encodes the Duffy-binding protein, which mediates P vivax’s invasion of erythrocytes. It was present in 5% of Papua Indonesia samples, 35% of western Cambodia samples, and 25% of western Thailand samples.
The third common CNV was a 37-kb duplication on chromosome 10 that includes pvmdr1, which has been associated with resistance to mefloquine and is homologous to the pfmdr1 amplification responsible for mefloquine resistance in P falciparum. This CNV was only present in samples from western Thailand.
The fourth common CNV was a 3-kb duplication on chromosome 14 that includes the gene PVX_101445. It was found only in Papua Indonesia samples.
“Our study shows that the strongest evidence of evolution is in Papua, Indonesia, where resistance of P vivax to chloroquine is now rampant,” said Ric Price, MD, of the University of Oxford in the UK.
“These data provide crucial information from which we can start to identify the mechanisms of drug resistance in P vivax.”
“We can see in the genome that drug resistance is a huge driver for evolution,” added Richard Pearson, PhD, of the Wellcome Trust Sanger Institute.
“Intriguingly, in some places, this process appears to be happening in response to drugs used primarily to treat a different malaria parasite, P falciparum. Although the exact cause isn’t known, this is a worrying sign that drug resistance is becoming deeply entrenched in the parasite population.”
The researchers said there are a few possible reasons why P vivax may be evolving to evade drugs used against P falciparum.
Many people carry mixed infections of both species of parasite, so, in treating one species, the other automatically gets exposed to the drug. Another culprit may be unsupervised drug use—where many people take the most readily available, rather than the most suitable, antimalarial drug.
Another finding from this study was that, when the researchers identified patients who were carrying multiple strains of parasite, the genomic data made it possible to determine how closely the different strains were related to one another.
“This means that we can now start to pull apart the genetic complexity of individual Plasmodium vivax infections and work out whether the parasites came from one or more mosquito bites,” Kwiatkowski said. “It provides a way of addressing fundamental questions about how P vivax is transmitted and how it persists within a community and, in particular, about the biology of relapsing infections.”

Plasmodium vivax
Image by Mae Melvin
Genomic research suggests the malaria parasite Plasmodium vivax is evolving rapidly to adapt to conditions in different geographic locations.
Researchers studied more than 200 parasite samples from across the Asia-Pacific region and found that P vivax has evolved differently in different areas.
The team identified substantial differences in the frequency of copy number variations (CNVs) in samples from western Thailand, western Cambodia, and Papua Indonesia.
They believe this is a result of the different antimalarial drugs used in these regions.
The researchers described this work in Nature Genetics.
“For so long, it’s not been possible to study P vivax genomes in detail, on a large-scale, but now we can, and we’re seeing the effect that drug use has on how parasites are evolving,” said study author Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in the UK.
He and his colleagues studied the genomes of 228 parasite samples, identifying the strains carried by each patient and revealing their infection history. Most samples came from Southeast Asia (Thailand, Cambodia, Vietnam, Laos, Myanmar, and Malaysia) and Oceania (Papua Indonesia and Papua New Guinea), but the team also studied samples from China, India, Sri Lanka, Brazil, and Madagascar.
The researchers performed detailed population genetic analyses using 148 samples from western Thailand, western Cambodia, and Papua Indonesia. This revealed CNVs in 9 regions of the core genome, and the frequency of the 4 most common CNVs varied greatly according to geographical location.
The first common CNV was a 9-kb deletion on chromosome 8 that includes the first 3 exons of a gene encoding a cytoadherence-linked asexual protein. The CNV was present in 73% of Papua Indonesia samples, 6% of western Cambodia samples, and 3% of western Thailand samples.
The second common CNV was a 7-kb duplication on chromosome 6 that encompasses pvdbp, the gene that encodes the Duffy-binding protein, which mediates P vivax’s invasion of erythrocytes. It was present in 5% of Papua Indonesia samples, 35% of western Cambodia samples, and 25% of western Thailand samples.
The third common CNV was a 37-kb duplication on chromosome 10 that includes pvmdr1, which has been associated with resistance to mefloquine and is homologous to the pfmdr1 amplification responsible for mefloquine resistance in P falciparum. This CNV was only present in samples from western Thailand.
The fourth common CNV was a 3-kb duplication on chromosome 14 that includes the gene PVX_101445. It was found only in Papua Indonesia samples.
“Our study shows that the strongest evidence of evolution is in Papua, Indonesia, where resistance of P vivax to chloroquine is now rampant,” said Ric Price, MD, of the University of Oxford in the UK.
“These data provide crucial information from which we can start to identify the mechanisms of drug resistance in P vivax.”
“We can see in the genome that drug resistance is a huge driver for evolution,” added Richard Pearson, PhD, of the Wellcome Trust Sanger Institute.
“Intriguingly, in some places, this process appears to be happening in response to drugs used primarily to treat a different malaria parasite, P falciparum. Although the exact cause isn’t known, this is a worrying sign that drug resistance is becoming deeply entrenched in the parasite population.”
The researchers said there are a few possible reasons why P vivax may be evolving to evade drugs used against P falciparum.
Many people carry mixed infections of both species of parasite, so, in treating one species, the other automatically gets exposed to the drug. Another culprit may be unsupervised drug use—where many people take the most readily available, rather than the most suitable, antimalarial drug.
Another finding from this study was that, when the researchers identified patients who were carrying multiple strains of parasite, the genomic data made it possible to determine how closely the different strains were related to one another.
“This means that we can now start to pull apart the genetic complexity of individual Plasmodium vivax infections and work out whether the parasites came from one or more mosquito bites,” Kwiatkowski said. “It provides a way of addressing fundamental questions about how P vivax is transmitted and how it persists within a community and, in particular, about the biology of relapsing infections.”
Immunotherapy drugs linked to rheumatic diseases

Photo by Bill Branson
Several case reports have suggested that cancer patients taking the immunotherapy drugs nivolumab and ipilimumab may have a higher-than-normal risk of developing rheumatic diseases.
Between 2012 and 2016, 13 patients at the Johns Hopkins Kimmel Cancer Center who were taking one or both drugs developed inflammatory arthritis or sicca syndrome, a set of autoimmune conditions causing dry eyes and mouth.
The cases were described in Annals of Rheumatic Diseases.
Nivolumab and ipilimumab are both designed to turn off the molecular “checkpoints” some cancers—including lymphoma—use to evade the immune system. When the drugs work, they allow the immune system to detect and attack tumor cells. However, they also turn up the activity of the immune system as a whole and can therefore trigger immune-related side effects.
Clinical trials of ipilimumab and nivolumab have indicated that the drugs confer an increased risk of inflammatory bowel diseases, lung inflammation, autoimmune thyroid disease, and pituitary gland inflammation.
However, those trials were designed primarily to determine efficacy against cancer and not to fully examine all features of rheumatologic side effects, said Laura C. Cappelli, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland.
With this in mind, she and her colleagues decided to take a closer look at 13 adults (older than 18) who were treated at the Johns Hopkins Kimmel Cancer Center and reported rheumatologic symptoms after their treatment with nivolumab and/or ipilimumab.
Eight patients were taking both ipilimumab and nivolumab, and 5 were taking 1 of the 2 drugs. They were receiving the drugs to treat melanoma (n=6), non-small-cell lung cancer (n=5), small-cell lung cancer (n=1), and renal cell carcinoma (n=1).
Nine of the patients developed inflammatory arthritis—4 with synovitis confirmed via imaging and 4 with inflammatory synovial fluid—and the remaining 4 patients were diagnosed with sicca syndrome. Other immune-related adverse events included pneumonitis, colitis, interstitial nephritis, and thyroiditis.
The researchers said this is the largest published case series showing a link between checkpoint inhibitors and rheumatic diseases.
The patients described in this case report make up about 1.3% of all patients treated with the drugs—singly or in combination—at The Johns Hopkins Hospital from 2012 to 2016. However, the researchers believe that rate is likely an underestimation of how common rheumatic diseases are in patients taking immune checkpoint inhibitors.
“We keep having referrals coming in from our oncologists as more patients are treated with these drugs,” said Clifton Bingham, MD, of the Johns Hopkins University School of Medicine.
“In particular, as more patients are treated with combinations of multiple immunotherapies, we expect the rate to go up.”
Dr Cappelli said she wants the case report to raise awareness among patients and clinicians that rheumatologic side effects may occur with checkpoint inhibitors.
“It is important when weighing the risk-benefit ratio of prescribing these drugs,” she said. “And it’s important for people to be on the lookout for symptoms so they can see a rheumatologist early in an effort to prevent or limit joint damage.”
Drs Cappelli and Bingham and their colleagues are planning further collaboration with Johns Hopkins oncologists to better track the incidence of rheumatic disease in patients taking immunotherapy drugs and determine whether any particular characteristics put cancer patients at higher risk of such complications.

Photo by Bill Branson
Several case reports have suggested that cancer patients taking the immunotherapy drugs nivolumab and ipilimumab may have a higher-than-normal risk of developing rheumatic diseases.
Between 2012 and 2016, 13 patients at the Johns Hopkins Kimmel Cancer Center who were taking one or both drugs developed inflammatory arthritis or sicca syndrome, a set of autoimmune conditions causing dry eyes and mouth.
The cases were described in Annals of Rheumatic Diseases.
Nivolumab and ipilimumab are both designed to turn off the molecular “checkpoints” some cancers—including lymphoma—use to evade the immune system. When the drugs work, they allow the immune system to detect and attack tumor cells. However, they also turn up the activity of the immune system as a whole and can therefore trigger immune-related side effects.
Clinical trials of ipilimumab and nivolumab have indicated that the drugs confer an increased risk of inflammatory bowel diseases, lung inflammation, autoimmune thyroid disease, and pituitary gland inflammation.
However, those trials were designed primarily to determine efficacy against cancer and not to fully examine all features of rheumatologic side effects, said Laura C. Cappelli, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland.
With this in mind, she and her colleagues decided to take a closer look at 13 adults (older than 18) who were treated at the Johns Hopkins Kimmel Cancer Center and reported rheumatologic symptoms after their treatment with nivolumab and/or ipilimumab.
Eight patients were taking both ipilimumab and nivolumab, and 5 were taking 1 of the 2 drugs. They were receiving the drugs to treat melanoma (n=6), non-small-cell lung cancer (n=5), small-cell lung cancer (n=1), and renal cell carcinoma (n=1).
Nine of the patients developed inflammatory arthritis—4 with synovitis confirmed via imaging and 4 with inflammatory synovial fluid—and the remaining 4 patients were diagnosed with sicca syndrome. Other immune-related adverse events included pneumonitis, colitis, interstitial nephritis, and thyroiditis.
The researchers said this is the largest published case series showing a link between checkpoint inhibitors and rheumatic diseases.
The patients described in this case report make up about 1.3% of all patients treated with the drugs—singly or in combination—at The Johns Hopkins Hospital from 2012 to 2016. However, the researchers believe that rate is likely an underestimation of how common rheumatic diseases are in patients taking immune checkpoint inhibitors.
“We keep having referrals coming in from our oncologists as more patients are treated with these drugs,” said Clifton Bingham, MD, of the Johns Hopkins University School of Medicine.
“In particular, as more patients are treated with combinations of multiple immunotherapies, we expect the rate to go up.”
Dr Cappelli said she wants the case report to raise awareness among patients and clinicians that rheumatologic side effects may occur with checkpoint inhibitors.
“It is important when weighing the risk-benefit ratio of prescribing these drugs,” she said. “And it’s important for people to be on the lookout for symptoms so they can see a rheumatologist early in an effort to prevent or limit joint damage.”
Drs Cappelli and Bingham and their colleagues are planning further collaboration with Johns Hopkins oncologists to better track the incidence of rheumatic disease in patients taking immunotherapy drugs and determine whether any particular characteristics put cancer patients at higher risk of such complications.

Photo by Bill Branson
Several case reports have suggested that cancer patients taking the immunotherapy drugs nivolumab and ipilimumab may have a higher-than-normal risk of developing rheumatic diseases.
Between 2012 and 2016, 13 patients at the Johns Hopkins Kimmel Cancer Center who were taking one or both drugs developed inflammatory arthritis or sicca syndrome, a set of autoimmune conditions causing dry eyes and mouth.
The cases were described in Annals of Rheumatic Diseases.
Nivolumab and ipilimumab are both designed to turn off the molecular “checkpoints” some cancers—including lymphoma—use to evade the immune system. When the drugs work, they allow the immune system to detect and attack tumor cells. However, they also turn up the activity of the immune system as a whole and can therefore trigger immune-related side effects.
Clinical trials of ipilimumab and nivolumab have indicated that the drugs confer an increased risk of inflammatory bowel diseases, lung inflammation, autoimmune thyroid disease, and pituitary gland inflammation.
However, those trials were designed primarily to determine efficacy against cancer and not to fully examine all features of rheumatologic side effects, said Laura C. Cappelli, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland.
With this in mind, she and her colleagues decided to take a closer look at 13 adults (older than 18) who were treated at the Johns Hopkins Kimmel Cancer Center and reported rheumatologic symptoms after their treatment with nivolumab and/or ipilimumab.
Eight patients were taking both ipilimumab and nivolumab, and 5 were taking 1 of the 2 drugs. They were receiving the drugs to treat melanoma (n=6), non-small-cell lung cancer (n=5), small-cell lung cancer (n=1), and renal cell carcinoma (n=1).
Nine of the patients developed inflammatory arthritis—4 with synovitis confirmed via imaging and 4 with inflammatory synovial fluid—and the remaining 4 patients were diagnosed with sicca syndrome. Other immune-related adverse events included pneumonitis, colitis, interstitial nephritis, and thyroiditis.
The researchers said this is the largest published case series showing a link between checkpoint inhibitors and rheumatic diseases.
The patients described in this case report make up about 1.3% of all patients treated with the drugs—singly or in combination—at The Johns Hopkins Hospital from 2012 to 2016. However, the researchers believe that rate is likely an underestimation of how common rheumatic diseases are in patients taking immune checkpoint inhibitors.
“We keep having referrals coming in from our oncologists as more patients are treated with these drugs,” said Clifton Bingham, MD, of the Johns Hopkins University School of Medicine.
“In particular, as more patients are treated with combinations of multiple immunotherapies, we expect the rate to go up.”
Dr Cappelli said she wants the case report to raise awareness among patients and clinicians that rheumatologic side effects may occur with checkpoint inhibitors.
“It is important when weighing the risk-benefit ratio of prescribing these drugs,” she said. “And it’s important for people to be on the lookout for symptoms so they can see a rheumatologist early in an effort to prevent or limit joint damage.”
Drs Cappelli and Bingham and their colleagues are planning further collaboration with Johns Hopkins oncologists to better track the incidence of rheumatic disease in patients taking immunotherapy drugs and determine whether any particular characteristics put cancer patients at higher risk of such complications.
HCPs may underestimate cancer risk from CT scans
Photo by Angela Mary Butler
Healthcare professionals (HCPs) may not be fully aware of a CT scan’s effect on lifetime malignancy risk, according to a study published in the Journal of Medical Imaging and Radiation Sciences.
Researchers surveyed a group of HCPs on radiation exposure from CT.
And although most of the respondents recognized that CT scans confer an increased risk of cancer, many underestimated the actual dose of radiation a person receives from a CT scan.
The survey was given to 308 HCPs—including physicians, radiologists, and technologists—in Saskatchewan, Canada.
Seventy-three percent of physicians, 97% of radiologists, and 76% of technologists correctly reported that there is an increased cancer risk from one abdominal-pelvic CT.
However, only 18% of physicians, 28% of radiologists, and 22% of technologists were able to correctly identify the dose in relation to chest X-rays.
In fact, 14% of physicians and 12% of technologists (but 0% of radiologists) “vastly” underestimated the dose as less than 10 chest X-ray equivalents, according to researchers.
The average radiation dose from an abdominal-pelvic CT is 10 millisieverts (mSv), compared to 0.02 mSv to 0.2 mSv from one chest X-ray, meaning that a radiation dose from a CT scan is equivalent to the dose from 100 to 250 chest radiographs.
“Underestimating radiation dose from a CT scan is more concerning than knowing the exact dose level, particularly when it is a vast underestimation, as this may lead to minimization of the risk estimate when considering a test,” said study author David Leswick, MD, of the University of Saskatchewan in Saskatoon.
“Although [cancer] risk from radiation dose levels in the range of medical imaging procedures is small, it is real, as evidenced from atomic bomb survivors and nuclear industry workers showing significantly increased risk of malignancy after exposure to doses in the range of diagnostic CT.”
“The risk of fatal malignancy may be as high as 1 in 1000 for a 10-mSv exposure. This risk is significant on a population basis, with up to 2% of cancers in the United States population possibly attributable to CT.”
Another aspect highlighted by the survey was some confusion regarding radiation exposure from MRIs and ultrasounds.
MRIs and ultrasounds do not employ ionizing radiation. However, 20% of physicians, 6% of radiologists, and 7% of technologists attributed radiation exposure to MRIs. Eleven percent of physicians, 0% of radiologists, and 7% of technologists believed an ultrasound used radiation.
“Belief that ionizing radiation is utilized by ultrasound and MRI is troubling, as it may result in underutilization of these imaging modalities because of unfounded radiation concerns,” Dr Leswick said.
“It is important for healthcare professionals (including referring physicians, radiologists, and technologists) to be aware of radiation dose levels and risks from imaging tests for several reasons, including the ability to weigh the risks and benefits of tests, counsel patients on relevant risks, optimize protocols to minimize radiation dose, and select appropriate protocols to minimize radiation dose.”

Photo by Angela Mary Butler
Healthcare professionals (HCPs) may not be fully aware of a CT scan’s effect on lifetime malignancy risk, according to a study published in the Journal of Medical Imaging and Radiation Sciences.
Researchers surveyed a group of HCPs on radiation exposure from CT.
And although most of the respondents recognized that CT scans confer an increased risk of cancer, many underestimated the actual dose of radiation a person receives from a CT scan.
The survey was given to 308 HCPs—including physicians, radiologists, and technologists—in Saskatchewan, Canada.
Seventy-three percent of physicians, 97% of radiologists, and 76% of technologists correctly reported that there is an increased cancer risk from one abdominal-pelvic CT.
However, only 18% of physicians, 28% of radiologists, and 22% of technologists were able to correctly identify the dose in relation to chest X-rays.
In fact, 14% of physicians and 12% of technologists (but 0% of radiologists) “vastly” underestimated the dose as less than 10 chest X-ray equivalents, according to researchers.
The average radiation dose from an abdominal-pelvic CT is 10 millisieverts (mSv), compared to 0.02 mSv to 0.2 mSv from one chest X-ray, meaning that a radiation dose from a CT scan is equivalent to the dose from 100 to 250 chest radiographs.
“Underestimating radiation dose from a CT scan is more concerning than knowing the exact dose level, particularly when it is a vast underestimation, as this may lead to minimization of the risk estimate when considering a test,” said study author David Leswick, MD, of the University of Saskatchewan in Saskatoon.
“Although [cancer] risk from radiation dose levels in the range of medical imaging procedures is small, it is real, as evidenced from atomic bomb survivors and nuclear industry workers showing significantly increased risk of malignancy after exposure to doses in the range of diagnostic CT.”
“The risk of fatal malignancy may be as high as 1 in 1000 for a 10-mSv exposure. This risk is significant on a population basis, with up to 2% of cancers in the United States population possibly attributable to CT.”
Another aspect highlighted by the survey was some confusion regarding radiation exposure from MRIs and ultrasounds.
MRIs and ultrasounds do not employ ionizing radiation. However, 20% of physicians, 6% of radiologists, and 7% of technologists attributed radiation exposure to MRIs. Eleven percent of physicians, 0% of radiologists, and 7% of technologists believed an ultrasound used radiation.
“Belief that ionizing radiation is utilized by ultrasound and MRI is troubling, as it may result in underutilization of these imaging modalities because of unfounded radiation concerns,” Dr Leswick said.
“It is important for healthcare professionals (including referring physicians, radiologists, and technologists) to be aware of radiation dose levels and risks from imaging tests for several reasons, including the ability to weigh the risks and benefits of tests, counsel patients on relevant risks, optimize protocols to minimize radiation dose, and select appropriate protocols to minimize radiation dose.”

Photo by Angela Mary Butler
Healthcare professionals (HCPs) may not be fully aware of a CT scan’s effect on lifetime malignancy risk, according to a study published in the Journal of Medical Imaging and Radiation Sciences.
Researchers surveyed a group of HCPs on radiation exposure from CT.
And although most of the respondents recognized that CT scans confer an increased risk of cancer, many underestimated the actual dose of radiation a person receives from a CT scan.
The survey was given to 308 HCPs—including physicians, radiologists, and technologists—in Saskatchewan, Canada.
Seventy-three percent of physicians, 97% of radiologists, and 76% of technologists correctly reported that there is an increased cancer risk from one abdominal-pelvic CT.
However, only 18% of physicians, 28% of radiologists, and 22% of technologists were able to correctly identify the dose in relation to chest X-rays.
In fact, 14% of physicians and 12% of technologists (but 0% of radiologists) “vastly” underestimated the dose as less than 10 chest X-ray equivalents, according to researchers.
The average radiation dose from an abdominal-pelvic CT is 10 millisieverts (mSv), compared to 0.02 mSv to 0.2 mSv from one chest X-ray, meaning that a radiation dose from a CT scan is equivalent to the dose from 100 to 250 chest radiographs.
“Underestimating radiation dose from a CT scan is more concerning than knowing the exact dose level, particularly when it is a vast underestimation, as this may lead to minimization of the risk estimate when considering a test,” said study author David Leswick, MD, of the University of Saskatchewan in Saskatoon.
“Although [cancer] risk from radiation dose levels in the range of medical imaging procedures is small, it is real, as evidenced from atomic bomb survivors and nuclear industry workers showing significantly increased risk of malignancy after exposure to doses in the range of diagnostic CT.”
“The risk of fatal malignancy may be as high as 1 in 1000 for a 10-mSv exposure. This risk is significant on a population basis, with up to 2% of cancers in the United States population possibly attributable to CT.”
Another aspect highlighted by the survey was some confusion regarding radiation exposure from MRIs and ultrasounds.
MRIs and ultrasounds do not employ ionizing radiation. However, 20% of physicians, 6% of radiologists, and 7% of technologists attributed radiation exposure to MRIs. Eleven percent of physicians, 0% of radiologists, and 7% of technologists believed an ultrasound used radiation.
“Belief that ionizing radiation is utilized by ultrasound and MRI is troubling, as it may result in underutilization of these imaging modalities because of unfounded radiation concerns,” Dr Leswick said.
“It is important for healthcare professionals (including referring physicians, radiologists, and technologists) to be aware of radiation dose levels and risks from imaging tests for several reasons, including the ability to weigh the risks and benefits of tests, counsel patients on relevant risks, optimize protocols to minimize radiation dose, and select appropriate protocols to minimize radiation dose.”