User login
FDA authorizes use of new Zika assay
The US Food and Drug Administration (FDA) has granted emergency use authorization (EUA) for a new assay designed to detect Zika virus infection.
The Aptima® Zika Virus assay is a molecular diagnostic tool that can be used to detect RNA from the Zika virus in human serum and plasma specimens.
The assay has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means the Aptima Zika Virus assay is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About the assay
The Aptima Zika Virus assay was developed by Hologic, Inc. The assay runs on the Hologic Panther® system, an integrated platform that fully automates all aspects of nucleic acid amplification testing.
The Aptima Zika Virus assay will be available for use in all 50 states, Puerto Rico, and US territories.
The assay is designed to be used in individuals meeting Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiologic criteria for which Zika virus testing may be indicated), by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988, 42 U.S.C. § 263a, to perform high complexity tests, or by similarly qualified non-US laboratories.
For more information about the Aptima Zika Virus assay, visit www.hologic.com/zika.
About Zika testing
The FDA has granted EUAs for 4 other tests designed to detect Zika virus:
- RealStar® Zika Virus RT-PCR Kit U.S. (altona Diagnostics GmbH)
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA, CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus. ![]()
The US Food and Drug Administration (FDA) has granted emergency use authorization (EUA) for a new assay designed to detect Zika virus infection.
The Aptima® Zika Virus assay is a molecular diagnostic tool that can be used to detect RNA from the Zika virus in human serum and plasma specimens.
The assay has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means the Aptima Zika Virus assay is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About the assay
The Aptima Zika Virus assay was developed by Hologic, Inc. The assay runs on the Hologic Panther® system, an integrated platform that fully automates all aspects of nucleic acid amplification testing.
The Aptima Zika Virus assay will be available for use in all 50 states, Puerto Rico, and US territories.
The assay is designed to be used in individuals meeting Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiologic criteria for which Zika virus testing may be indicated), by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988, 42 U.S.C. § 263a, to perform high complexity tests, or by similarly qualified non-US laboratories.
For more information about the Aptima Zika Virus assay, visit www.hologic.com/zika.
About Zika testing
The FDA has granted EUAs for 4 other tests designed to detect Zika virus:
- RealStar® Zika Virus RT-PCR Kit U.S. (altona Diagnostics GmbH)
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA, CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus. ![]()
The US Food and Drug Administration (FDA) has granted emergency use authorization (EUA) for a new assay designed to detect Zika virus infection.
The Aptima® Zika Virus assay is a molecular diagnostic tool that can be used to detect RNA from the Zika virus in human serum and plasma specimens.
The assay has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means the Aptima Zika Virus assay is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About the assay
The Aptima Zika Virus assay was developed by Hologic, Inc. The assay runs on the Hologic Panther® system, an integrated platform that fully automates all aspects of nucleic acid amplification testing.
The Aptima Zika Virus assay will be available for use in all 50 states, Puerto Rico, and US territories.
The assay is designed to be used in individuals meeting Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiologic criteria for which Zika virus testing may be indicated), by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988, 42 U.S.C. § 263a, to perform high complexity tests, or by similarly qualified non-US laboratories.
For more information about the Aptima Zika Virus assay, visit www.hologic.com/zika.
About Zika testing
The FDA has granted EUAs for 4 other tests designed to detect Zika virus:
- RealStar® Zika Virus RT-PCR Kit U.S. (altona Diagnostics GmbH)
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA, CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus. ![]()
Antiplatelet therapy could treat Alzheimer’s disease
Image by Andre E.X. Brown
Antiplatelet drugs can significantly reduce amyloid plaques in cerebral vessels of transgenic mice with Alzheimer’s disease, according to new research.
The study revealed a mechanism for direct involvement of platelets in the progression of Alzheimer’s disease, and investigators believe this could be of great importance for the treatment of Alzheimer’s patients.
Alzheimer’s disease is characterized by the formation of amyloid aggregates and deposits of amyloid in the brain. The amyloid deposits damage the structure and function of nerve tissue in the brain and lead to the loss of neuronal cells and cognitive capability.
Amyloid deposits in Alzheimer’s disease occur not only in the brain parenchyma, but also in blood vessels in the brain. The current study, published in Science Signaling, deals with the vascular form of the disease.
Previous research demonstrated that platelets attach to amyloid deposits in the vessel wall, which leads to ongoing platelet activation in mice. The platelets then form a hemostatic plug, which occludes vessels in the brain and leads to insufficient perfusion of the surrounding tissue.
Investigators have now determined that the protein amyloid-ß binds to a specific integrin on the platelet surface that is important for the aggregation of platelets.
This binding induces the release of adenosine diphosphate and the chaperone protein clusterin and supports the formation of amyloid plaques.
In cell culture experiments, the investigators analyzed platelets from 5 patients with Glanzmann’s thrombasthenia, a hereditary defect of platelet activation, and found no amyloid plaques.
The team then treated Alzheimer’s transgenic mice with the antiplatelet agent clopidogrel. The mice exhibited reduced platelet activation and significantly reduced amyloid plaque formation, which improved the perfusion of the brain during the 3-month treatment with the drug.
The investigators suggest that antiplatelet therapy may alleviate fibril formation in cerebral vessels of Alzheimer’s disease patients. ![]()

Image by Andre E.X. Brown
Antiplatelet drugs can significantly reduce amyloid plaques in cerebral vessels of transgenic mice with Alzheimer’s disease, according to new research.
The study revealed a mechanism for direct involvement of platelets in the progression of Alzheimer’s disease, and investigators believe this could be of great importance for the treatment of Alzheimer’s patients.
Alzheimer’s disease is characterized by the formation of amyloid aggregates and deposits of amyloid in the brain. The amyloid deposits damage the structure and function of nerve tissue in the brain and lead to the loss of neuronal cells and cognitive capability.
Amyloid deposits in Alzheimer’s disease occur not only in the brain parenchyma, but also in blood vessels in the brain. The current study, published in Science Signaling, deals with the vascular form of the disease.
Previous research demonstrated that platelets attach to amyloid deposits in the vessel wall, which leads to ongoing platelet activation in mice. The platelets then form a hemostatic plug, which occludes vessels in the brain and leads to insufficient perfusion of the surrounding tissue.
Investigators have now determined that the protein amyloid-ß binds to a specific integrin on the platelet surface that is important for the aggregation of platelets.
This binding induces the release of adenosine diphosphate and the chaperone protein clusterin and supports the formation of amyloid plaques.
In cell culture experiments, the investigators analyzed platelets from 5 patients with Glanzmann’s thrombasthenia, a hereditary defect of platelet activation, and found no amyloid plaques.
The team then treated Alzheimer’s transgenic mice with the antiplatelet agent clopidogrel. The mice exhibited reduced platelet activation and significantly reduced amyloid plaque formation, which improved the perfusion of the brain during the 3-month treatment with the drug.
The investigators suggest that antiplatelet therapy may alleviate fibril formation in cerebral vessels of Alzheimer’s disease patients. ![]()

Image by Andre E.X. Brown
Antiplatelet drugs can significantly reduce amyloid plaques in cerebral vessels of transgenic mice with Alzheimer’s disease, according to new research.
The study revealed a mechanism for direct involvement of platelets in the progression of Alzheimer’s disease, and investigators believe this could be of great importance for the treatment of Alzheimer’s patients.
Alzheimer’s disease is characterized by the formation of amyloid aggregates and deposits of amyloid in the brain. The amyloid deposits damage the structure and function of nerve tissue in the brain and lead to the loss of neuronal cells and cognitive capability.
Amyloid deposits in Alzheimer’s disease occur not only in the brain parenchyma, but also in blood vessels in the brain. The current study, published in Science Signaling, deals with the vascular form of the disease.
Previous research demonstrated that platelets attach to amyloid deposits in the vessel wall, which leads to ongoing platelet activation in mice. The platelets then form a hemostatic plug, which occludes vessels in the brain and leads to insufficient perfusion of the surrounding tissue.
Investigators have now determined that the protein amyloid-ß binds to a specific integrin on the platelet surface that is important for the aggregation of platelets.
This binding induces the release of adenosine diphosphate and the chaperone protein clusterin and supports the formation of amyloid plaques.
In cell culture experiments, the investigators analyzed platelets from 5 patients with Glanzmann’s thrombasthenia, a hereditary defect of platelet activation, and found no amyloid plaques.
The team then treated Alzheimer’s transgenic mice with the antiplatelet agent clopidogrel. The mice exhibited reduced platelet activation and significantly reduced amyloid plaque formation, which improved the perfusion of the brain during the 3-month treatment with the drug.
The investigators suggest that antiplatelet therapy may alleviate fibril formation in cerebral vessels of Alzheimer’s disease patients. ![]()
Late sepsis death not explained by pre-existing conditions

Photo courtesy of the CDC
A new study sheds light on whether an increased risk of death in the 30 days to 2 years after contracting sepsis is caused by sepsis itself, or because of pre-existing health conditions the patients had before acquiring the complication.
Using detailed survey data and medical records of more than 30,000 older Americans, the researchers conducted a propensity matched cohort study to investigate the phenomenon of late death after sepsis.
Late death refers to deaths that take place months to years after the acute infection has resolved.
"We know sicker patients are more likely to develop sepsis," lead author Hallie Prescott, MD, of the University of Michigan in Ann Arbor, said. "And that made us wonder: Perhaps those previous health conditions are driving the risk of late death after sepsis?"
So the investigators compared 960 patients aged 65 or older who were admitted to the hospital with sepsis to 3 control groups: 777 adults not currently hospitalized, 788 patients hospitalized with non-sepsis infections, and 504 patients admitted with acute sterile inflammatory conditions.
All patients had participated in the US Health and Retirement Study, a longitudinal survey of 37,000 adults aged over 50 in 23,000 households. The survey is considered broadly representative of the older US population.
The current study included Medicare beneficiaries who had participated in at least one survey between 1998 and 2008.
The main outcome measure was late mortality 31 days to 2 years after sepsis and odds of death at various intervals.
Results
In the sepsis cohort, the mean age was 79, 54% were women, 81% were white, and 12% were nursing home residents.
There were no significant differences among the cohorts in terms of demographics, socioeconomic characteristics, baseline health status, or recent healthcare use.
The sepsis cohort had a 25.4% mortality rate at 30 days, 35.3% at 90 days, 41.3% at 180 days, 48.5% at 1 year, and 56.5% at 2 years.
Over 40% of sepsis patients who survived 30 days after their hospitalization died in the next 2 years. If sepsis patients survived to a year, the adjusted 2-year mortality was 16.0% versus 10.7% for controls not in the hospital.
When investigators compared these mortality rates to matched patients who were not hospitalized, they found that those with sepsis experienced a 22.1% absolute increase in late mortality. This translated into a 2.2-fold relative increase in late mortality.
And when they compared the sepsis patients to patients hospitalized for non-sepsis infections, the investigators determined that sepsis patients experienced a 10.4% absolute increase in late mortality. This translated into a 1.3-fold relative increase in late mortality.
Finally, they compared the sepsis patients to those patients hospitalized for sterile inflammatory conditions and found that the sepsis patients who survived to 31 days experienced a 16.2% absolute increase in late mortality. This translated into 1.6-fold relative increase.
The investigators said this high rate of late mortality could not be explained by the patients’ age, socio-demographics, or their pre-sepsis health status.
"Rather, we found that, compared to the group of adults not in the hospital,” Dr Prescott said, “1 in 5 patients who survived sepsis had a late death that was not explained by their baseline characteristics.”
The investigators said this suggests that late mortality after sepsis could be more amendable to intervention than previously thought.
The investigators published their findings in BMJ. ![]()

Photo courtesy of the CDC
A new study sheds light on whether an increased risk of death in the 30 days to 2 years after contracting sepsis is caused by sepsis itself, or because of pre-existing health conditions the patients had before acquiring the complication.
Using detailed survey data and medical records of more than 30,000 older Americans, the researchers conducted a propensity matched cohort study to investigate the phenomenon of late death after sepsis.
Late death refers to deaths that take place months to years after the acute infection has resolved.
"We know sicker patients are more likely to develop sepsis," lead author Hallie Prescott, MD, of the University of Michigan in Ann Arbor, said. "And that made us wonder: Perhaps those previous health conditions are driving the risk of late death after sepsis?"
So the investigators compared 960 patients aged 65 or older who were admitted to the hospital with sepsis to 3 control groups: 777 adults not currently hospitalized, 788 patients hospitalized with non-sepsis infections, and 504 patients admitted with acute sterile inflammatory conditions.
All patients had participated in the US Health and Retirement Study, a longitudinal survey of 37,000 adults aged over 50 in 23,000 households. The survey is considered broadly representative of the older US population.
The current study included Medicare beneficiaries who had participated in at least one survey between 1998 and 2008.
The main outcome measure was late mortality 31 days to 2 years after sepsis and odds of death at various intervals.
Results
In the sepsis cohort, the mean age was 79, 54% were women, 81% were white, and 12% were nursing home residents.
There were no significant differences among the cohorts in terms of demographics, socioeconomic characteristics, baseline health status, or recent healthcare use.
The sepsis cohort had a 25.4% mortality rate at 30 days, 35.3% at 90 days, 41.3% at 180 days, 48.5% at 1 year, and 56.5% at 2 years.
Over 40% of sepsis patients who survived 30 days after their hospitalization died in the next 2 years. If sepsis patients survived to a year, the adjusted 2-year mortality was 16.0% versus 10.7% for controls not in the hospital.
When investigators compared these mortality rates to matched patients who were not hospitalized, they found that those with sepsis experienced a 22.1% absolute increase in late mortality. This translated into a 2.2-fold relative increase in late mortality.
And when they compared the sepsis patients to patients hospitalized for non-sepsis infections, the investigators determined that sepsis patients experienced a 10.4% absolute increase in late mortality. This translated into a 1.3-fold relative increase in late mortality.
Finally, they compared the sepsis patients to those patients hospitalized for sterile inflammatory conditions and found that the sepsis patients who survived to 31 days experienced a 16.2% absolute increase in late mortality. This translated into 1.6-fold relative increase.
The investigators said this high rate of late mortality could not be explained by the patients’ age, socio-demographics, or their pre-sepsis health status.
"Rather, we found that, compared to the group of adults not in the hospital,” Dr Prescott said, “1 in 5 patients who survived sepsis had a late death that was not explained by their baseline characteristics.”
The investigators said this suggests that late mortality after sepsis could be more amendable to intervention than previously thought.
The investigators published their findings in BMJ. ![]()

Photo courtesy of the CDC
A new study sheds light on whether an increased risk of death in the 30 days to 2 years after contracting sepsis is caused by sepsis itself, or because of pre-existing health conditions the patients had before acquiring the complication.
Using detailed survey data and medical records of more than 30,000 older Americans, the researchers conducted a propensity matched cohort study to investigate the phenomenon of late death after sepsis.
Late death refers to deaths that take place months to years after the acute infection has resolved.
"We know sicker patients are more likely to develop sepsis," lead author Hallie Prescott, MD, of the University of Michigan in Ann Arbor, said. "And that made us wonder: Perhaps those previous health conditions are driving the risk of late death after sepsis?"
So the investigators compared 960 patients aged 65 or older who were admitted to the hospital with sepsis to 3 control groups: 777 adults not currently hospitalized, 788 patients hospitalized with non-sepsis infections, and 504 patients admitted with acute sterile inflammatory conditions.
All patients had participated in the US Health and Retirement Study, a longitudinal survey of 37,000 adults aged over 50 in 23,000 households. The survey is considered broadly representative of the older US population.
The current study included Medicare beneficiaries who had participated in at least one survey between 1998 and 2008.
The main outcome measure was late mortality 31 days to 2 years after sepsis and odds of death at various intervals.
Results
In the sepsis cohort, the mean age was 79, 54% were women, 81% were white, and 12% were nursing home residents.
There were no significant differences among the cohorts in terms of demographics, socioeconomic characteristics, baseline health status, or recent healthcare use.
The sepsis cohort had a 25.4% mortality rate at 30 days, 35.3% at 90 days, 41.3% at 180 days, 48.5% at 1 year, and 56.5% at 2 years.
Over 40% of sepsis patients who survived 30 days after their hospitalization died in the next 2 years. If sepsis patients survived to a year, the adjusted 2-year mortality was 16.0% versus 10.7% for controls not in the hospital.
When investigators compared these mortality rates to matched patients who were not hospitalized, they found that those with sepsis experienced a 22.1% absolute increase in late mortality. This translated into a 2.2-fold relative increase in late mortality.
And when they compared the sepsis patients to patients hospitalized for non-sepsis infections, the investigators determined that sepsis patients experienced a 10.4% absolute increase in late mortality. This translated into a 1.3-fold relative increase in late mortality.
Finally, they compared the sepsis patients to those patients hospitalized for sterile inflammatory conditions and found that the sepsis patients who survived to 31 days experienced a 16.2% absolute increase in late mortality. This translated into 1.6-fold relative increase.
The investigators said this high rate of late mortality could not be explained by the patients’ age, socio-demographics, or their pre-sepsis health status.
"Rather, we found that, compared to the group of adults not in the hospital,” Dr Prescott said, “1 in 5 patients who survived sepsis had a late death that was not explained by their baseline characteristics.”
The investigators said this suggests that late mortality after sepsis could be more amendable to intervention than previously thought.
The investigators published their findings in BMJ. ![]()
Sodium influx may be key to killing Plasmodium parasites
invading an RBC
Credit: St Jude Children's
Research Hospital
Two new anti-malaria drug candidates with different mechanisms of action—a pyrazoleamide and a spiroindolone—promote an influx of sodium ions into Plasmodium parasites that have invaded red blood cells and multiply there.
Within minutes, the increase in sodium kills the parasites, investigators believe, by changing its outer membrane and promoting division before its genome has been replicated.
Akhil Vaidya, PhD, of Drexel University College of Medicine in Philadelphia, and members of the research team published details of their findings in PLOS Pathogens.
The Plasmodium plasma membrane contains very low levels of cholesterol, which is a major lipid component of most other cell membranes.
Saponin, a detergent that can dissolve cholesterol-containing membranes, dissolves red blood cells infected by Plasmodium and releases intact parasites into the bloodstream. The detergent is unable to destroy the parasites because their membranes have low cholesterol content.
However, when researchers exposed the parasite cell membranes to the 2 drugs, they became permeable by saponin. The researchers deemed this to be a function of the increased amount of cholesterol incorporated into the parasite membrane.
“We believe that the cholesterol makes the parasite rigid, and then the parasite can no longer pass through very small spaces in the bloodstream,” Dr Vaidya said. The parasite cannot continue its lifecycle if it cannot enter red blood cells.
Researchers also discovered that when drug exposure is short, the changes in membrane composition are reversible. The parasites regain their resistance to saponin most likely because the additional membrane cholesterol washes off.
After 2 hours of treatment with either drug, many of the parasites had fragmented nuclei and interior membranes. Researchers did not observe any sign of multiplication of the parasite genome, which is necessary to create daughter cells and precedes other cell division events.
The researchers were surprised by the findings. They had assumed that the spiroindolone, KAE609 (cipargamin), which is being investigated in clinical trials, killed parasites through a different mechanism.
The investigators maintain that by understanding exactly how new drug candidates stop malaria, they will learn more about the parasite’s vulnerabilities and be able to determine the origin of drug resistance as soon as it arises.
“We want to find the best ways to keep new drugs effective as long as we can,” Dr Vaidya said.
This study was funded by National Institutes of Health Grant R01-AI98413 and Medicines for Malaria Venture Grant MMV/08/0027. ![]()

invading an RBC
Credit: St Jude Children's
Research Hospital
Two new anti-malaria drug candidates with different mechanisms of action—a pyrazoleamide and a spiroindolone—promote an influx of sodium ions into Plasmodium parasites that have invaded red blood cells and multiply there.
Within minutes, the increase in sodium kills the parasites, investigators believe, by changing its outer membrane and promoting division before its genome has been replicated.
Akhil Vaidya, PhD, of Drexel University College of Medicine in Philadelphia, and members of the research team published details of their findings in PLOS Pathogens.
The Plasmodium plasma membrane contains very low levels of cholesterol, which is a major lipid component of most other cell membranes.
Saponin, a detergent that can dissolve cholesterol-containing membranes, dissolves red blood cells infected by Plasmodium and releases intact parasites into the bloodstream. The detergent is unable to destroy the parasites because their membranes have low cholesterol content.
However, when researchers exposed the parasite cell membranes to the 2 drugs, they became permeable by saponin. The researchers deemed this to be a function of the increased amount of cholesterol incorporated into the parasite membrane.
“We believe that the cholesterol makes the parasite rigid, and then the parasite can no longer pass through very small spaces in the bloodstream,” Dr Vaidya said. The parasite cannot continue its lifecycle if it cannot enter red blood cells.
Researchers also discovered that when drug exposure is short, the changes in membrane composition are reversible. The parasites regain their resistance to saponin most likely because the additional membrane cholesterol washes off.
After 2 hours of treatment with either drug, many of the parasites had fragmented nuclei and interior membranes. Researchers did not observe any sign of multiplication of the parasite genome, which is necessary to create daughter cells and precedes other cell division events.
The researchers were surprised by the findings. They had assumed that the spiroindolone, KAE609 (cipargamin), which is being investigated in clinical trials, killed parasites through a different mechanism.
The investigators maintain that by understanding exactly how new drug candidates stop malaria, they will learn more about the parasite’s vulnerabilities and be able to determine the origin of drug resistance as soon as it arises.
“We want to find the best ways to keep new drugs effective as long as we can,” Dr Vaidya said.
This study was funded by National Institutes of Health Grant R01-AI98413 and Medicines for Malaria Venture Grant MMV/08/0027. ![]()

invading an RBC
Credit: St Jude Children's
Research Hospital
Two new anti-malaria drug candidates with different mechanisms of action—a pyrazoleamide and a spiroindolone—promote an influx of sodium ions into Plasmodium parasites that have invaded red blood cells and multiply there.
Within minutes, the increase in sodium kills the parasites, investigators believe, by changing its outer membrane and promoting division before its genome has been replicated.
Akhil Vaidya, PhD, of Drexel University College of Medicine in Philadelphia, and members of the research team published details of their findings in PLOS Pathogens.
The Plasmodium plasma membrane contains very low levels of cholesterol, which is a major lipid component of most other cell membranes.
Saponin, a detergent that can dissolve cholesterol-containing membranes, dissolves red blood cells infected by Plasmodium and releases intact parasites into the bloodstream. The detergent is unable to destroy the parasites because their membranes have low cholesterol content.
However, when researchers exposed the parasite cell membranes to the 2 drugs, they became permeable by saponin. The researchers deemed this to be a function of the increased amount of cholesterol incorporated into the parasite membrane.
“We believe that the cholesterol makes the parasite rigid, and then the parasite can no longer pass through very small spaces in the bloodstream,” Dr Vaidya said. The parasite cannot continue its lifecycle if it cannot enter red blood cells.
Researchers also discovered that when drug exposure is short, the changes in membrane composition are reversible. The parasites regain their resistance to saponin most likely because the additional membrane cholesterol washes off.
After 2 hours of treatment with either drug, many of the parasites had fragmented nuclei and interior membranes. Researchers did not observe any sign of multiplication of the parasite genome, which is necessary to create daughter cells and precedes other cell division events.
The researchers were surprised by the findings. They had assumed that the spiroindolone, KAE609 (cipargamin), which is being investigated in clinical trials, killed parasites through a different mechanism.
The investigators maintain that by understanding exactly how new drug candidates stop malaria, they will learn more about the parasite’s vulnerabilities and be able to determine the origin of drug resistance as soon as it arises.
“We want to find the best ways to keep new drugs effective as long as we can,” Dr Vaidya said.
This study was funded by National Institutes of Health Grant R01-AI98413 and Medicines for Malaria Venture Grant MMV/08/0027. ![]()
Team develops new approach to programming T cells
Using mouse models, researchers have developed a new cellular programming approach to create alloreactive T cells they say eliminate leukemic cells without causing graft-versus-host disease (GVHD).
They created the T cells using the donor key immune cell. When used in allogeneic hematopoietic stem cell transplantation and anti-leukemia therapy, the new approach reduced the toxicities that cause GVHD while preserving the anti-leukemia activity of the immune cell.
“This approach will be useful in the future when developing novel methods for immunotherapy,” said Yi Zhang, MD, PhD, of Temple University in Philadelphia, Pennsylvania.
Dr Zhang and colleagues took murine bone marrow using Flt3 ligand and Toll-like receptor agonists to produce δ-like ligand 4-positive dendritic cells (Dll4hiDCs). When the dendritic cells were stimulated, CD4+ naïve T cells underwent effector differentiation and produced high levels of IFN-γ and IL-17 in vitro.
The team then transferred the allogeneic Dll4hiDC-induced T cells into the mice. The cells did not induce severe GVHD and preserved anti-leukemic activity, “significantly improving the survival of leukemic mice undergoing allogeneic HSCT,” they said.
They noted that the IFN-γ was important for Dll4hiDC programming in reducing the GVHD toxicities of alloreactive T cells. When the researchers transferred unstimulated T cells into mice, 5 of 8 mice died from GVHD and 3 of 8 died with tumor. Those that received Dll4hiDC-induced T cells did not develop GVHD.
They also emphasized that this platform does not require transfection with viral vectors, which has limitations of safety and efficiency.
“This system will not only be useful for reducing GvHD,” Dr Zhang said, “but can also be used in the identification of T cells for the improvement of other types of immunotherapy for advanced cancer.”
The team published this research in Blood. ![]()

Using mouse models, researchers have developed a new cellular programming approach to create alloreactive T cells they say eliminate leukemic cells without causing graft-versus-host disease (GVHD).
They created the T cells using the donor key immune cell. When used in allogeneic hematopoietic stem cell transplantation and anti-leukemia therapy, the new approach reduced the toxicities that cause GVHD while preserving the anti-leukemia activity of the immune cell.
“This approach will be useful in the future when developing novel methods for immunotherapy,” said Yi Zhang, MD, PhD, of Temple University in Philadelphia, Pennsylvania.
Dr Zhang and colleagues took murine bone marrow using Flt3 ligand and Toll-like receptor agonists to produce δ-like ligand 4-positive dendritic cells (Dll4hiDCs). When the dendritic cells were stimulated, CD4+ naïve T cells underwent effector differentiation and produced high levels of IFN-γ and IL-17 in vitro.
The team then transferred the allogeneic Dll4hiDC-induced T cells into the mice. The cells did not induce severe GVHD and preserved anti-leukemic activity, “significantly improving the survival of leukemic mice undergoing allogeneic HSCT,” they said.
They noted that the IFN-γ was important for Dll4hiDC programming in reducing the GVHD toxicities of alloreactive T cells. When the researchers transferred unstimulated T cells into mice, 5 of 8 mice died from GVHD and 3 of 8 died with tumor. Those that received Dll4hiDC-induced T cells did not develop GVHD.
They also emphasized that this platform does not require transfection with viral vectors, which has limitations of safety and efficiency.
“This system will not only be useful for reducing GvHD,” Dr Zhang said, “but can also be used in the identification of T cells for the improvement of other types of immunotherapy for advanced cancer.”
The team published this research in Blood. ![]()

Using mouse models, researchers have developed a new cellular programming approach to create alloreactive T cells they say eliminate leukemic cells without causing graft-versus-host disease (GVHD).
They created the T cells using the donor key immune cell. When used in allogeneic hematopoietic stem cell transplantation and anti-leukemia therapy, the new approach reduced the toxicities that cause GVHD while preserving the anti-leukemia activity of the immune cell.
“This approach will be useful in the future when developing novel methods for immunotherapy,” said Yi Zhang, MD, PhD, of Temple University in Philadelphia, Pennsylvania.
Dr Zhang and colleagues took murine bone marrow using Flt3 ligand and Toll-like receptor agonists to produce δ-like ligand 4-positive dendritic cells (Dll4hiDCs). When the dendritic cells were stimulated, CD4+ naïve T cells underwent effector differentiation and produced high levels of IFN-γ and IL-17 in vitro.
The team then transferred the allogeneic Dll4hiDC-induced T cells into the mice. The cells did not induce severe GVHD and preserved anti-leukemic activity, “significantly improving the survival of leukemic mice undergoing allogeneic HSCT,” they said.
They noted that the IFN-γ was important for Dll4hiDC programming in reducing the GVHD toxicities of alloreactive T cells. When the researchers transferred unstimulated T cells into mice, 5 of 8 mice died from GVHD and 3 of 8 died with tumor. Those that received Dll4hiDC-induced T cells did not develop GVHD.
They also emphasized that this platform does not require transfection with viral vectors, which has limitations of safety and efficiency.
“This system will not only be useful for reducing GvHD,” Dr Zhang said, “but can also be used in the identification of T cells for the improvement of other types of immunotherapy for advanced cancer.”
The team published this research in Blood. ![]()
Two compounds show promise against Zika virus

Photo courtesy of
Muhammad Mahdi Karim
Two compounds have shown activity against the Zika virus, according to Biotech Biotron, a company that develops compounds to fight viral diseases such as HIV and hepatitis C. The two compounds from its library killed the Zika virus in vitro, as determined by an independent USA laboratory facility.
“These early results are encouraging,” Michelle Miller, PhD, of Biotron, said. “Identification of these active compounds in our library is a starting point for designing potent drugs against Zika.”
At present, there is no approved vaccine or treatment for Zika virus, whose common symptoms include fever, rash, joint pain, and conjunctivitis.
While the symptoms are generally mild, Zika infection during pregnancy has been associated with microcephaly and other severe brain defects in the newborn.
In addition, Zika infection may be associated with an increased risk of Guillain-Barré syndrome, which is being investigated by the Centers for Disease Control and Prevention.
Biotron is planning to carry out more tests on the Zika virus to determine whether the compounds are likely to be safe and effective in humans.
The Zika virus is primarily spread by infected mosquitoes. But exposure to an infected person’s blood or other body fluids may also result in transmission. ![]()

Photo courtesy of
Muhammad Mahdi Karim
Two compounds have shown activity against the Zika virus, according to Biotech Biotron, a company that develops compounds to fight viral diseases such as HIV and hepatitis C. The two compounds from its library killed the Zika virus in vitro, as determined by an independent USA laboratory facility.
“These early results are encouraging,” Michelle Miller, PhD, of Biotron, said. “Identification of these active compounds in our library is a starting point for designing potent drugs against Zika.”
At present, there is no approved vaccine or treatment for Zika virus, whose common symptoms include fever, rash, joint pain, and conjunctivitis.
While the symptoms are generally mild, Zika infection during pregnancy has been associated with microcephaly and other severe brain defects in the newborn.
In addition, Zika infection may be associated with an increased risk of Guillain-Barré syndrome, which is being investigated by the Centers for Disease Control and Prevention.
Biotron is planning to carry out more tests on the Zika virus to determine whether the compounds are likely to be safe and effective in humans.
The Zika virus is primarily spread by infected mosquitoes. But exposure to an infected person’s blood or other body fluids may also result in transmission. ![]()

Photo courtesy of
Muhammad Mahdi Karim
Two compounds have shown activity against the Zika virus, according to Biotech Biotron, a company that develops compounds to fight viral diseases such as HIV and hepatitis C. The two compounds from its library killed the Zika virus in vitro, as determined by an independent USA laboratory facility.
“These early results are encouraging,” Michelle Miller, PhD, of Biotron, said. “Identification of these active compounds in our library is a starting point for designing potent drugs against Zika.”
At present, there is no approved vaccine or treatment for Zika virus, whose common symptoms include fever, rash, joint pain, and conjunctivitis.
While the symptoms are generally mild, Zika infection during pregnancy has been associated with microcephaly and other severe brain defects in the newborn.
In addition, Zika infection may be associated with an increased risk of Guillain-Barré syndrome, which is being investigated by the Centers for Disease Control and Prevention.
Biotron is planning to carry out more tests on the Zika virus to determine whether the compounds are likely to be safe and effective in humans.
The Zika virus is primarily spread by infected mosquitoes. But exposure to an infected person’s blood or other body fluids may also result in transmission. ![]()
Negative RT-PCR result doesn’t exclude Zika infection
Photo by Jeremy L. Grisham
The Centers for Disease Control and Prevention (CDC) has issued an interim guidance on how to interpret results of the Zika virus antibody test.
Over the past few weeks, the FDA has authorized the use of new Zika tests, including RealStar® Zika Virus RT-PCR Kit U.S., Zika Virus RNA Qualitative Real-Time RT-PCR test, and the Trioplex Real-time RT-PCR Assay.
The CDC is now updating its guidance, since a negative real time reverse transcription-polymerase chain reaction (rRT-PCR) test does not necessarily rule out Zika infection.
In these cases, the CDC recommends immunoglobulin (Ig) M and neutralizing antibody testing, which can identify additional recent Zika virus infections.
The Zika antibody test, however, is difficult to interpret because of cross-reactivity with other flaviviruses. Zika is a mosquito-borne flavivirus that is closely related to dengue, West Nile, Japanese encephalitis, and yellow fever viruses.
The cross-reactivity can preclude identification of the specific infecting virus, particularly if a person was previously infected with or vaccinated against a related flavivirus. And appropriate clinical management is dependent upon proper identification of the virus.
If IgM test results are positive, equivocal, or inconclusive, the CDC recommends performing a plaque reduction neutralization test (PRNT) to confirm the diagnosis.
However, in people who have been previously infected with or vaccinated against a related flavivirus, even a 4-fold higher titer by PRNT may not be able to discriminate between anti-Zika virus antibodies and cross-reacting antibodies.
Therefore, the CDC now recommends an even more conservative approach to interpreting PRNT results to reduce the possibility of missing the diagnosis of either Zika or dengue virus infection.
The US Food and Drug Administration issued in February an Emergency Use Authorization for the CDC Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA) for antibody testing.
It is used to detect Zika virus IgM antibodies in serum or cerebrospinal fluid from people with suspect Zika virus disease.
Presumptive positive results should be confirmed with PRNT against Zika, dengue, and other flaviviruses.
Equivocal and inconclusive results that are not resolved by re-testing should also have PRNT performed to rule out false-positive results.
For more information on interpretation of the Zika virus antibody test results, see the CDC’s Interim Guidance published as part of the Morbidity and Mortality Weekly Report for 31 May 2016. ![]()

Photo by Jeremy L. Grisham
The Centers for Disease Control and Prevention (CDC) has issued an interim guidance on how to interpret results of the Zika virus antibody test.
Over the past few weeks, the FDA has authorized the use of new Zika tests, including RealStar® Zika Virus RT-PCR Kit U.S., Zika Virus RNA Qualitative Real-Time RT-PCR test, and the Trioplex Real-time RT-PCR Assay.
The CDC is now updating its guidance, since a negative real time reverse transcription-polymerase chain reaction (rRT-PCR) test does not necessarily rule out Zika infection.
In these cases, the CDC recommends immunoglobulin (Ig) M and neutralizing antibody testing, which can identify additional recent Zika virus infections.
The Zika antibody test, however, is difficult to interpret because of cross-reactivity with other flaviviruses. Zika is a mosquito-borne flavivirus that is closely related to dengue, West Nile, Japanese encephalitis, and yellow fever viruses.
The cross-reactivity can preclude identification of the specific infecting virus, particularly if a person was previously infected with or vaccinated against a related flavivirus. And appropriate clinical management is dependent upon proper identification of the virus.
If IgM test results are positive, equivocal, or inconclusive, the CDC recommends performing a plaque reduction neutralization test (PRNT) to confirm the diagnosis.
However, in people who have been previously infected with or vaccinated against a related flavivirus, even a 4-fold higher titer by PRNT may not be able to discriminate between anti-Zika virus antibodies and cross-reacting antibodies.
Therefore, the CDC now recommends an even more conservative approach to interpreting PRNT results to reduce the possibility of missing the diagnosis of either Zika or dengue virus infection.
The US Food and Drug Administration issued in February an Emergency Use Authorization for the CDC Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA) for antibody testing.
It is used to detect Zika virus IgM antibodies in serum or cerebrospinal fluid from people with suspect Zika virus disease.
Presumptive positive results should be confirmed with PRNT against Zika, dengue, and other flaviviruses.
Equivocal and inconclusive results that are not resolved by re-testing should also have PRNT performed to rule out false-positive results.
For more information on interpretation of the Zika virus antibody test results, see the CDC’s Interim Guidance published as part of the Morbidity and Mortality Weekly Report for 31 May 2016. ![]()

Photo by Jeremy L. Grisham
The Centers for Disease Control and Prevention (CDC) has issued an interim guidance on how to interpret results of the Zika virus antibody test.
Over the past few weeks, the FDA has authorized the use of new Zika tests, including RealStar® Zika Virus RT-PCR Kit U.S., Zika Virus RNA Qualitative Real-Time RT-PCR test, and the Trioplex Real-time RT-PCR Assay.
The CDC is now updating its guidance, since a negative real time reverse transcription-polymerase chain reaction (rRT-PCR) test does not necessarily rule out Zika infection.
In these cases, the CDC recommends immunoglobulin (Ig) M and neutralizing antibody testing, which can identify additional recent Zika virus infections.
The Zika antibody test, however, is difficult to interpret because of cross-reactivity with other flaviviruses. Zika is a mosquito-borne flavivirus that is closely related to dengue, West Nile, Japanese encephalitis, and yellow fever viruses.
The cross-reactivity can preclude identification of the specific infecting virus, particularly if a person was previously infected with or vaccinated against a related flavivirus. And appropriate clinical management is dependent upon proper identification of the virus.
If IgM test results are positive, equivocal, or inconclusive, the CDC recommends performing a plaque reduction neutralization test (PRNT) to confirm the diagnosis.
However, in people who have been previously infected with or vaccinated against a related flavivirus, even a 4-fold higher titer by PRNT may not be able to discriminate between anti-Zika virus antibodies and cross-reacting antibodies.
Therefore, the CDC now recommends an even more conservative approach to interpreting PRNT results to reduce the possibility of missing the diagnosis of either Zika or dengue virus infection.
The US Food and Drug Administration issued in February an Emergency Use Authorization for the CDC Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA) for antibody testing.
It is used to detect Zika virus IgM antibodies in serum or cerebrospinal fluid from people with suspect Zika virus disease.
Presumptive positive results should be confirmed with PRNT against Zika, dengue, and other flaviviruses.
Equivocal and inconclusive results that are not resolved by re-testing should also have PRNT performed to rule out false-positive results.
For more information on interpretation of the Zika virus antibody test results, see the CDC’s Interim Guidance published as part of the Morbidity and Mortality Weekly Report for 31 May 2016.
Why patients don’t report possible cancer symptoms

Photo courtesy of NIH
Worrying about wasting their doctor’s time is stopping people from reporting symptoms that might be related to cancer, according to a small study published in the British Journal of General Practice.
The goal of the study was to determine why some people are more likely than others to worry about wasting a general practitioner’s (GP’s) time and delay reporting possible cancer symptoms.
“People worrying about wasting their doctor’s time is one of the challenges we need to tackle when thinking about trying to diagnose cancer earlier,” said study author Katriina Whitaker, PhD, of the University of Surrey in the UK.
“We need to get to the root of the problem and find out why people are feeling worried. Not a lot of work has been done on this so far. Our study draws attention to some reasons patients put off going to their GP to check out possible cancer symptoms.”
For this study, Dr Whitaker and her colleagues conducted interviews with subjects in London, South East England, and North West England.
The subjects were recruited from a sample of 2042 adults, age 50 and older, who completed a survey that included a list of “cancer alarm symptoms.”
Ultimately, the researchers interviewed 62 subjects who had reported symptoms at baseline, were still present at the 3-month follow-up, and had agreed to be contacted.
The interviews revealed a few reasons why subjects were hesitant to report symptoms to their GP.
Some subjects felt that long waiting times for appointments indicated GPs were very busy, so they shouldn’t bother making an appointment unless symptoms seemed very serious.
Other subjects felt that seeking help when their symptoms did not seem serious—ie, persistent, worsening, or life-threatening—was a waste of a doctor’s time.
Still other subjects were hesitant to seek help because their doctors had been dismissive about symptoms in the past.
On the other hand, subjects who reported positive interactions with GPs or good relationships with them were less worried about time-wasting.
And other subjects weren’t worried about wasting their doctor’s time because they think of GPs as fulfilling a service financed by taxpayers.
“We’ve all had times where we’ve wondered if we should go to see a GP, but getting unusual or persistent changes checked out is really important,” said Julie Sharp, head of health and patient information at Cancer Research UK, which funded this study.
“Worrying about wasting a GP’s time should not put people off. Doctors are there to help spot cancer symptoms early when treatment is more likely to be successful, and delaying a visit could save up bigger problems for later. So if you’ve noticed anything that isn’t normal for you, make an appointment to see your doctor.”

Photo courtesy of NIH
Worrying about wasting their doctor’s time is stopping people from reporting symptoms that might be related to cancer, according to a small study published in the British Journal of General Practice.
The goal of the study was to determine why some people are more likely than others to worry about wasting a general practitioner’s (GP’s) time and delay reporting possible cancer symptoms.
“People worrying about wasting their doctor’s time is one of the challenges we need to tackle when thinking about trying to diagnose cancer earlier,” said study author Katriina Whitaker, PhD, of the University of Surrey in the UK.
“We need to get to the root of the problem and find out why people are feeling worried. Not a lot of work has been done on this so far. Our study draws attention to some reasons patients put off going to their GP to check out possible cancer symptoms.”
For this study, Dr Whitaker and her colleagues conducted interviews with subjects in London, South East England, and North West England.
The subjects were recruited from a sample of 2042 adults, age 50 and older, who completed a survey that included a list of “cancer alarm symptoms.”
Ultimately, the researchers interviewed 62 subjects who had reported symptoms at baseline, were still present at the 3-month follow-up, and had agreed to be contacted.
The interviews revealed a few reasons why subjects were hesitant to report symptoms to their GP.
Some subjects felt that long waiting times for appointments indicated GPs were very busy, so they shouldn’t bother making an appointment unless symptoms seemed very serious.
Other subjects felt that seeking help when their symptoms did not seem serious—ie, persistent, worsening, or life-threatening—was a waste of a doctor’s time.
Still other subjects were hesitant to seek help because their doctors had been dismissive about symptoms in the past.
On the other hand, subjects who reported positive interactions with GPs or good relationships with them were less worried about time-wasting.
And other subjects weren’t worried about wasting their doctor’s time because they think of GPs as fulfilling a service financed by taxpayers.
“We’ve all had times where we’ve wondered if we should go to see a GP, but getting unusual or persistent changes checked out is really important,” said Julie Sharp, head of health and patient information at Cancer Research UK, which funded this study.
“Worrying about wasting a GP’s time should not put people off. Doctors are there to help spot cancer symptoms early when treatment is more likely to be successful, and delaying a visit could save up bigger problems for later. So if you’ve noticed anything that isn’t normal for you, make an appointment to see your doctor.”

Photo courtesy of NIH
Worrying about wasting their doctor’s time is stopping people from reporting symptoms that might be related to cancer, according to a small study published in the British Journal of General Practice.
The goal of the study was to determine why some people are more likely than others to worry about wasting a general practitioner’s (GP’s) time and delay reporting possible cancer symptoms.
“People worrying about wasting their doctor’s time is one of the challenges we need to tackle when thinking about trying to diagnose cancer earlier,” said study author Katriina Whitaker, PhD, of the University of Surrey in the UK.
“We need to get to the root of the problem and find out why people are feeling worried. Not a lot of work has been done on this so far. Our study draws attention to some reasons patients put off going to their GP to check out possible cancer symptoms.”
For this study, Dr Whitaker and her colleagues conducted interviews with subjects in London, South East England, and North West England.
The subjects were recruited from a sample of 2042 adults, age 50 and older, who completed a survey that included a list of “cancer alarm symptoms.”
Ultimately, the researchers interviewed 62 subjects who had reported symptoms at baseline, were still present at the 3-month follow-up, and had agreed to be contacted.
The interviews revealed a few reasons why subjects were hesitant to report symptoms to their GP.
Some subjects felt that long waiting times for appointments indicated GPs were very busy, so they shouldn’t bother making an appointment unless symptoms seemed very serious.
Other subjects felt that seeking help when their symptoms did not seem serious—ie, persistent, worsening, or life-threatening—was a waste of a doctor’s time.
Still other subjects were hesitant to seek help because their doctors had been dismissive about symptoms in the past.
On the other hand, subjects who reported positive interactions with GPs or good relationships with them were less worried about time-wasting.
And other subjects weren’t worried about wasting their doctor’s time because they think of GPs as fulfilling a service financed by taxpayers.
“We’ve all had times where we’ve wondered if we should go to see a GP, but getting unusual or persistent changes checked out is really important,” said Julie Sharp, head of health and patient information at Cancer Research UK, which funded this study.
“Worrying about wasting a GP’s time should not put people off. Doctors are there to help spot cancer symptoms early when treatment is more likely to be successful, and delaying a visit could save up bigger problems for later. So if you’ve noticed anything that isn’t normal for you, make an appointment to see your doctor.”
Proteins may be targets for malaria vaccines
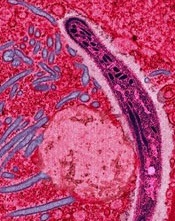
Image courtesy of Ute Frevert
and Margaret Shear
A study published in PLOS Pathogens has revealed proteins that may be viable targets for malaria vaccines.
Investigators identified 42 proteins that can be found on the surface of Plasmodium falciparum sporozoites and could be targeted by vaccines.
However, the team also found evidence to suggest that 2 other surface proteins should not be targeted, as they may be able to evade the immune system.
“We used a method that we developed in a previous paper to identify which proteins of the malaria parasite, Plasmodium falciparum, might be visible to the human immune system on the outside of the parasite and therefore are good potential targets for the development of new malaria vaccines,” said study author Scott E. Lindner, PhD, of Pennsylvania State University in University Park.
“Current experimental malaria vaccines target single proteins and do not provide the level of protection necessary to prevent the spread of the disease. Our new list of potential vaccine targets will allow the development of more effective vaccines that target several proteins on the surface of the parasite.”
To identify these targets, Dr Lindner and his colleagues collected malaria sporozoites from the salivary glands of thousands of infected mosquitoes.
The investigators then marked proteins on the surface of the sporozoites with a chemical label that could not cross through the outer membrane of the parasite. The team identified and characterized the labeled proteins using mass spectrometry.
“We focused on the transmission stage of the parasite because, at this point in an infection, the number of parasites is low, and if we can design effective vaccines for this stage, we can stop the progress of the disease before it causes symptoms,” Dr Lindner said. “Once the parasites are in the liver, they can hide from our immune system by residing inside of liver cells.”
Based on multiple replications of their experiments, the investigators identified 42 proteins that are highly likely to be exposed on the surface of the parasite and are therefore potential targets for vaccines.
The team noted that many of the proteins they identified had been thought to be located exclusively on the inside of the parasite. They suggest that these proteins may become exposed as the parasite moves from the site of a mosquito bite toward the liver.
“Malaria is still one of the great global health issues today, with hundreds of millions of new infections and half a million deaths each year, most of which occur in children under the age of 5,” Dr Lindner said.
“The parasite quickly and efficiently develops resistance to the drugs that we use to treat the disease, so what’s really needed to make eradication of malaria possible is a better vaccine. Our research provides an experimentally validated list of protein targets that could be used to develop new, more effective malaria vaccines.”
The investigators also discovered that 2 surface proteins—CSP and TRAP—are glycosylated in sporozoites, which changes the way the proteins are recognized by the immune system.
The team believes this discovery will affect the way future vaccines are designed as well.
“Our goal was to identify proteins that are present on the surface of sporozoites in hopes of finding targets for new vaccines,” said study author Kristian E. Swearingen, PhD, of the Institute for Systems Biology in Seattle, Washington.
“In addition to the potential new targets we’ve found, we’re also excited about the discovery that 2 of the major sporozoite surface proteins are glycosylated. The presence of sugars on these proteins almost certainly affects the way they are recognized by antibodies, something that will need to be factored in for future vaccine efforts based on these proteins.”

Image courtesy of Ute Frevert
and Margaret Shear
A study published in PLOS Pathogens has revealed proteins that may be viable targets for malaria vaccines.
Investigators identified 42 proteins that can be found on the surface of Plasmodium falciparum sporozoites and could be targeted by vaccines.
However, the team also found evidence to suggest that 2 other surface proteins should not be targeted, as they may be able to evade the immune system.
“We used a method that we developed in a previous paper to identify which proteins of the malaria parasite, Plasmodium falciparum, might be visible to the human immune system on the outside of the parasite and therefore are good potential targets for the development of new malaria vaccines,” said study author Scott E. Lindner, PhD, of Pennsylvania State University in University Park.
“Current experimental malaria vaccines target single proteins and do not provide the level of protection necessary to prevent the spread of the disease. Our new list of potential vaccine targets will allow the development of more effective vaccines that target several proteins on the surface of the parasite.”
To identify these targets, Dr Lindner and his colleagues collected malaria sporozoites from the salivary glands of thousands of infected mosquitoes.
The investigators then marked proteins on the surface of the sporozoites with a chemical label that could not cross through the outer membrane of the parasite. The team identified and characterized the labeled proteins using mass spectrometry.
“We focused on the transmission stage of the parasite because, at this point in an infection, the number of parasites is low, and if we can design effective vaccines for this stage, we can stop the progress of the disease before it causes symptoms,” Dr Lindner said. “Once the parasites are in the liver, they can hide from our immune system by residing inside of liver cells.”
Based on multiple replications of their experiments, the investigators identified 42 proteins that are highly likely to be exposed on the surface of the parasite and are therefore potential targets for vaccines.
The team noted that many of the proteins they identified had been thought to be located exclusively on the inside of the parasite. They suggest that these proteins may become exposed as the parasite moves from the site of a mosquito bite toward the liver.
“Malaria is still one of the great global health issues today, with hundreds of millions of new infections and half a million deaths each year, most of which occur in children under the age of 5,” Dr Lindner said.
“The parasite quickly and efficiently develops resistance to the drugs that we use to treat the disease, so what’s really needed to make eradication of malaria possible is a better vaccine. Our research provides an experimentally validated list of protein targets that could be used to develop new, more effective malaria vaccines.”
The investigators also discovered that 2 surface proteins—CSP and TRAP—are glycosylated in sporozoites, which changes the way the proteins are recognized by the immune system.
The team believes this discovery will affect the way future vaccines are designed as well.
“Our goal was to identify proteins that are present on the surface of sporozoites in hopes of finding targets for new vaccines,” said study author Kristian E. Swearingen, PhD, of the Institute for Systems Biology in Seattle, Washington.
“In addition to the potential new targets we’ve found, we’re also excited about the discovery that 2 of the major sporozoite surface proteins are glycosylated. The presence of sugars on these proteins almost certainly affects the way they are recognized by antibodies, something that will need to be factored in for future vaccine efforts based on these proteins.”

Image courtesy of Ute Frevert
and Margaret Shear
A study published in PLOS Pathogens has revealed proteins that may be viable targets for malaria vaccines.
Investigators identified 42 proteins that can be found on the surface of Plasmodium falciparum sporozoites and could be targeted by vaccines.
However, the team also found evidence to suggest that 2 other surface proteins should not be targeted, as they may be able to evade the immune system.
“We used a method that we developed in a previous paper to identify which proteins of the malaria parasite, Plasmodium falciparum, might be visible to the human immune system on the outside of the parasite and therefore are good potential targets for the development of new malaria vaccines,” said study author Scott E. Lindner, PhD, of Pennsylvania State University in University Park.
“Current experimental malaria vaccines target single proteins and do not provide the level of protection necessary to prevent the spread of the disease. Our new list of potential vaccine targets will allow the development of more effective vaccines that target several proteins on the surface of the parasite.”
To identify these targets, Dr Lindner and his colleagues collected malaria sporozoites from the salivary glands of thousands of infected mosquitoes.
The investigators then marked proteins on the surface of the sporozoites with a chemical label that could not cross through the outer membrane of the parasite. The team identified and characterized the labeled proteins using mass spectrometry.
“We focused on the transmission stage of the parasite because, at this point in an infection, the number of parasites is low, and if we can design effective vaccines for this stage, we can stop the progress of the disease before it causes symptoms,” Dr Lindner said. “Once the parasites are in the liver, they can hide from our immune system by residing inside of liver cells.”
Based on multiple replications of their experiments, the investigators identified 42 proteins that are highly likely to be exposed on the surface of the parasite and are therefore potential targets for vaccines.
The team noted that many of the proteins they identified had been thought to be located exclusively on the inside of the parasite. They suggest that these proteins may become exposed as the parasite moves from the site of a mosquito bite toward the liver.
“Malaria is still one of the great global health issues today, with hundreds of millions of new infections and half a million deaths each year, most of which occur in children under the age of 5,” Dr Lindner said.
“The parasite quickly and efficiently develops resistance to the drugs that we use to treat the disease, so what’s really needed to make eradication of malaria possible is a better vaccine. Our research provides an experimentally validated list of protein targets that could be used to develop new, more effective malaria vaccines.”
The investigators also discovered that 2 surface proteins—CSP and TRAP—are glycosylated in sporozoites, which changes the way the proteins are recognized by the immune system.
The team believes this discovery will affect the way future vaccines are designed as well.
“Our goal was to identify proteins that are present on the surface of sporozoites in hopes of finding targets for new vaccines,” said study author Kristian E. Swearingen, PhD, of the Institute for Systems Biology in Seattle, Washington.
“In addition to the potential new targets we’ve found, we’re also excited about the discovery that 2 of the major sporozoite surface proteins are glycosylated. The presence of sugars on these proteins almost certainly affects the way they are recognized by antibodies, something that will need to be factored in for future vaccine efforts based on these proteins.”
FDA authorizes use of new Zika test

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has granted Emergency Use Authorization (EUA) for another test designed to detect Zika virus infection.
The RealStar® Zika Virus RT-PCR Kit U.S. is a product of altona Diagnostics GmbH.
Under the EUA, the test can be used as a molecular diagnostic tool for the in vitro qualitative detection of RNA from the Zika virus in human serum or urine (collected alongside a patient-matched serum specimen).
The RealStar® Zika Virus RT-PCR Kit U.S. is intended for use in individuals meeting US Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika virus transmission at the time of travel).
The test is authorized for use only under the EUA in CLIA-certified High Complexity Laboratories in the US or by similarly qualified non-US laboratories, by clinical laboratory personnel specifically trained in the techniques of real-time PCR and in vitro diagnostic procedures.
The RealStar® Zika Virus RT-PCR Kit U.S. is authorized for a workflow consisting of nucleic acid extraction using the QIAamp® Viral RNA Mini Kit (QIAGEN), followed by the amplification and detection of Zika-virus-specific RNA using the RealStar® Zika Virus RT-PCR Kit U.S. on an ABI Prism® 7500 SDS/Fast SDS (Applied Biosystems), CFX96™ Real-Time PCR Detection System or CFX96™ Deep Well Real-Time PCR Detection System (both from BIO-RAD), LightCycler® 480 Instrument II (Roche), Rotor-Gene® 6000 (Corbett Research), or Rotor-Gene® Q 5/6 plex/MDxPlatform (QIAGEN).
About the EUA
The RealStar® Zika Virus RT-PCR Kit U.S. has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The RealStar® Zika Virus RT-PCR Kit U.S. is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About Zika testing
The FDA has granted EUAs for 3 other tests designed to detect Zika virus:
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus.

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has granted Emergency Use Authorization (EUA) for another test designed to detect Zika virus infection.
The RealStar® Zika Virus RT-PCR Kit U.S. is a product of altona Diagnostics GmbH.
Under the EUA, the test can be used as a molecular diagnostic tool for the in vitro qualitative detection of RNA from the Zika virus in human serum or urine (collected alongside a patient-matched serum specimen).
The RealStar® Zika Virus RT-PCR Kit U.S. is intended for use in individuals meeting US Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika virus transmission at the time of travel).
The test is authorized for use only under the EUA in CLIA-certified High Complexity Laboratories in the US or by similarly qualified non-US laboratories, by clinical laboratory personnel specifically trained in the techniques of real-time PCR and in vitro diagnostic procedures.
The RealStar® Zika Virus RT-PCR Kit U.S. is authorized for a workflow consisting of nucleic acid extraction using the QIAamp® Viral RNA Mini Kit (QIAGEN), followed by the amplification and detection of Zika-virus-specific RNA using the RealStar® Zika Virus RT-PCR Kit U.S. on an ABI Prism® 7500 SDS/Fast SDS (Applied Biosystems), CFX96™ Real-Time PCR Detection System or CFX96™ Deep Well Real-Time PCR Detection System (both from BIO-RAD), LightCycler® 480 Instrument II (Roche), Rotor-Gene® 6000 (Corbett Research), or Rotor-Gene® Q 5/6 plex/MDxPlatform (QIAGEN).
About the EUA
The RealStar® Zika Virus RT-PCR Kit U.S. has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The RealStar® Zika Virus RT-PCR Kit U.S. is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About Zika testing
The FDA has granted EUAs for 3 other tests designed to detect Zika virus:
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus.

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has granted Emergency Use Authorization (EUA) for another test designed to detect Zika virus infection.
The RealStar® Zika Virus RT-PCR Kit U.S. is a product of altona Diagnostics GmbH.
Under the EUA, the test can be used as a molecular diagnostic tool for the in vitro qualitative detection of RNA from the Zika virus in human serum or urine (collected alongside a patient-matched serum specimen).
The RealStar® Zika Virus RT-PCR Kit U.S. is intended for use in individuals meeting US Centers for Disease Control and Prevention (CDC) Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika virus transmission at the time of travel).
The test is authorized for use only under the EUA in CLIA-certified High Complexity Laboratories in the US or by similarly qualified non-US laboratories, by clinical laboratory personnel specifically trained in the techniques of real-time PCR and in vitro diagnostic procedures.
The RealStar® Zika Virus RT-PCR Kit U.S. is authorized for a workflow consisting of nucleic acid extraction using the QIAamp® Viral RNA Mini Kit (QIAGEN), followed by the amplification and detection of Zika-virus-specific RNA using the RealStar® Zika Virus RT-PCR Kit U.S. on an ABI Prism® 7500 SDS/Fast SDS (Applied Biosystems), CFX96™ Real-Time PCR Detection System or CFX96™ Deep Well Real-Time PCR Detection System (both from BIO-RAD), LightCycler® 480 Instrument II (Roche), Rotor-Gene® 6000 (Corbett Research), or Rotor-Gene® Q 5/6 plex/MDxPlatform (QIAGEN).
About the EUA
The RealStar® Zika Virus RT-PCR Kit U.S. has not been FDA-cleared or approved. An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The RealStar® Zika Virus RT-PCR Kit U.S. is only authorized as long as circumstances exist to justify the authorization of the emergency use of in vitro diagnostics for the detection of Zika virus under section 564(b)(1) of the Federal Food, Drug & Cosmetic Act, 21 U.S.C.§360bbb-3(b)(1), unless the authorization is terminated or revoked sooner.
About Zika testing
The FDA has granted EUAs for 3 other tests designed to detect Zika virus:
- Zika Virus RNA Qualitative Real-Time RT-PCR test (Focus Diagnostics)
- Trioplex Real-time RT-PCR Assay (CDC)
- Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (CDC).
The FDA has also authorized use of the cobas® Zika test (Roche) to screen blood donations for Zika virus. The test may be used under an investigational new drug application for screening donated blood in areas with active mosquito-borne transmission of the virus.