User login
Blood culture panel cleared by FDA
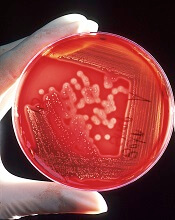
Staphylococcus infection
Photo by Bill Branson
The US Food and Drug Administration (FDA) has granted 510(k) clearance for a blood culture panel that detects sepsis caused by methicillin-resistant Staphylococcus aureus (MRSA) and other Staphylococcus species.
The Staph ID/R Blood Culture Panel is a product of Great Basin Scientific, Inc.
It is an automated, DNA multiplex assay used to identify Staphylococcus species directly from positive blood cultures in about 2 hours.
The panel also detects the mecA gene, a drug-resistance marker that confers resistance to methicillin and other beta-lactams and creates MRSA.
In addition, the Staph ID/R Blood Culture Panel identifies coagulase-negative staphylococci.
According to the US Centers for Disease Control and Prevention, 20% to 50% of all positive blood cultures are likely false positives due to contamination caused by coagulase-negative staphylococci, many of which are part of the normal flora of human skin and are not dangerous.
The Staph ID/R Blood Culture Panel is run on the Great Basin Analyzer. The company says the assay requires less than a minute of hands-on time and no results interpretation due to electronic results reporting. ![]()

Staphylococcus infection
Photo by Bill Branson
The US Food and Drug Administration (FDA) has granted 510(k) clearance for a blood culture panel that detects sepsis caused by methicillin-resistant Staphylococcus aureus (MRSA) and other Staphylococcus species.
The Staph ID/R Blood Culture Panel is a product of Great Basin Scientific, Inc.
It is an automated, DNA multiplex assay used to identify Staphylococcus species directly from positive blood cultures in about 2 hours.
The panel also detects the mecA gene, a drug-resistance marker that confers resistance to methicillin and other beta-lactams and creates MRSA.
In addition, the Staph ID/R Blood Culture Panel identifies coagulase-negative staphylococci.
According to the US Centers for Disease Control and Prevention, 20% to 50% of all positive blood cultures are likely false positives due to contamination caused by coagulase-negative staphylococci, many of which are part of the normal flora of human skin and are not dangerous.
The Staph ID/R Blood Culture Panel is run on the Great Basin Analyzer. The company says the assay requires less than a minute of hands-on time and no results interpretation due to electronic results reporting. ![]()

Staphylococcus infection
Photo by Bill Branson
The US Food and Drug Administration (FDA) has granted 510(k) clearance for a blood culture panel that detects sepsis caused by methicillin-resistant Staphylococcus aureus (MRSA) and other Staphylococcus species.
The Staph ID/R Blood Culture Panel is a product of Great Basin Scientific, Inc.
It is an automated, DNA multiplex assay used to identify Staphylococcus species directly from positive blood cultures in about 2 hours.
The panel also detects the mecA gene, a drug-resistance marker that confers resistance to methicillin and other beta-lactams and creates MRSA.
In addition, the Staph ID/R Blood Culture Panel identifies coagulase-negative staphylococci.
According to the US Centers for Disease Control and Prevention, 20% to 50% of all positive blood cultures are likely false positives due to contamination caused by coagulase-negative staphylococci, many of which are part of the normal flora of human skin and are not dangerous.
The Staph ID/R Blood Culture Panel is run on the Great Basin Analyzer. The company says the assay requires less than a minute of hands-on time and no results interpretation due to electronic results reporting. ![]()
Accuracy of blood test results varies

Photo by Graham Colm
A comparison of commercially available blood tests has revealed more variability than expected, according to researchers.
The group compared basic blood tests run by commercial laboratories and found the testing service, type of test, and time of collection all influenced the accuracy of results.
Given that these tests can be used for disease diagnosis or to determine whether a patient’s medication is working, the researchers said this study highlights the importance of knowing the accuracy and variability of blood test results.
“While most of the variability we found was within clinically accepted ranges, there were several cases where inaccurate results would have led to incorrect medical decisions,” said Joel Dudley, PhD, of the Icahn School of Medicine at Mount Sinai in New York, New York.
“We hope this study will inspire the biomedical community to take a critical look at all testing variables to ensure that lab results are as robust and reproducible as possible.”
Dr Dudley and his colleagues described this study in the Journal of Clinical Investigation.
The researchers collected peripheral blood samples from 60 healthy adults at 4 separate time points within a 6.5-hour window. The samples were collected in Phoenix, Arizona, at an ambulatory clinic and at retail outlets with point-of-care services.
The team collected 14 samples per subject and used those samples to compare 22 common clinical lab tests conducted at 3 commercial labs. One lab, Theranos, offered blood tests obtained from a finger prick, and the other 2, Quest and LabCorp, required standard venipuncture draws.
More than half of the test results showed significant differences between test providers. Of the 22 tests, 15 (68%) showed significant variability between labs (P<0.002).
Triglyceride levels and red blood cell counts were among the most consistent results, while white blood cell counts and overall cholesterol levels were among the most variable.
Test results from Theranos were flagged by Theranos as abnormal 1.6 times more often than tests from LabCorp or Quest (P<0.0001). The percentages for measurements outside their normal range were 8.3% for LabCorp, 7.5% for Quest, and 12.2% for Theranos.
In addition, the researchers noted that, although they controlled subjects’ eating and physical activity, data from blood samples collected earlier in the day were sometimes significantly different from samples taken from the same subjects later in the day.
There were significant difference between measurements collected at time points 1 and 2 vs time points 3 and 4 for 13 of the 22 tests (P<0.002).
“These testing disparities occurred despite rigorous laboratory certification and proficiency standards designed to ensure consistency,” said study author Eric Schadt, PhD, of Mount Sinai.
“Our results suggest the need for greater transparency in lab technologies and procedures, as well as a much more thorough investigation of biological mechanisms that may contribute to more dynamic levels than we currently understand.” ![]()

Photo by Graham Colm
A comparison of commercially available blood tests has revealed more variability than expected, according to researchers.
The group compared basic blood tests run by commercial laboratories and found the testing service, type of test, and time of collection all influenced the accuracy of results.
Given that these tests can be used for disease diagnosis or to determine whether a patient’s medication is working, the researchers said this study highlights the importance of knowing the accuracy and variability of blood test results.
“While most of the variability we found was within clinically accepted ranges, there were several cases where inaccurate results would have led to incorrect medical decisions,” said Joel Dudley, PhD, of the Icahn School of Medicine at Mount Sinai in New York, New York.
“We hope this study will inspire the biomedical community to take a critical look at all testing variables to ensure that lab results are as robust and reproducible as possible.”
Dr Dudley and his colleagues described this study in the Journal of Clinical Investigation.
The researchers collected peripheral blood samples from 60 healthy adults at 4 separate time points within a 6.5-hour window. The samples were collected in Phoenix, Arizona, at an ambulatory clinic and at retail outlets with point-of-care services.
The team collected 14 samples per subject and used those samples to compare 22 common clinical lab tests conducted at 3 commercial labs. One lab, Theranos, offered blood tests obtained from a finger prick, and the other 2, Quest and LabCorp, required standard venipuncture draws.
More than half of the test results showed significant differences between test providers. Of the 22 tests, 15 (68%) showed significant variability between labs (P<0.002).
Triglyceride levels and red blood cell counts were among the most consistent results, while white blood cell counts and overall cholesterol levels were among the most variable.
Test results from Theranos were flagged by Theranos as abnormal 1.6 times more often than tests from LabCorp or Quest (P<0.0001). The percentages for measurements outside their normal range were 8.3% for LabCorp, 7.5% for Quest, and 12.2% for Theranos.
In addition, the researchers noted that, although they controlled subjects’ eating and physical activity, data from blood samples collected earlier in the day were sometimes significantly different from samples taken from the same subjects later in the day.
There were significant difference between measurements collected at time points 1 and 2 vs time points 3 and 4 for 13 of the 22 tests (P<0.002).
“These testing disparities occurred despite rigorous laboratory certification and proficiency standards designed to ensure consistency,” said study author Eric Schadt, PhD, of Mount Sinai.
“Our results suggest the need for greater transparency in lab technologies and procedures, as well as a much more thorough investigation of biological mechanisms that may contribute to more dynamic levels than we currently understand.” ![]()

Photo by Graham Colm
A comparison of commercially available blood tests has revealed more variability than expected, according to researchers.
The group compared basic blood tests run by commercial laboratories and found the testing service, type of test, and time of collection all influenced the accuracy of results.
Given that these tests can be used for disease diagnosis or to determine whether a patient’s medication is working, the researchers said this study highlights the importance of knowing the accuracy and variability of blood test results.
“While most of the variability we found was within clinically accepted ranges, there were several cases where inaccurate results would have led to incorrect medical decisions,” said Joel Dudley, PhD, of the Icahn School of Medicine at Mount Sinai in New York, New York.
“We hope this study will inspire the biomedical community to take a critical look at all testing variables to ensure that lab results are as robust and reproducible as possible.”
Dr Dudley and his colleagues described this study in the Journal of Clinical Investigation.
The researchers collected peripheral blood samples from 60 healthy adults at 4 separate time points within a 6.5-hour window. The samples were collected in Phoenix, Arizona, at an ambulatory clinic and at retail outlets with point-of-care services.
The team collected 14 samples per subject and used those samples to compare 22 common clinical lab tests conducted at 3 commercial labs. One lab, Theranos, offered blood tests obtained from a finger prick, and the other 2, Quest and LabCorp, required standard venipuncture draws.
More than half of the test results showed significant differences between test providers. Of the 22 tests, 15 (68%) showed significant variability between labs (P<0.002).
Triglyceride levels and red blood cell counts were among the most consistent results, while white blood cell counts and overall cholesterol levels were among the most variable.
Test results from Theranos were flagged by Theranos as abnormal 1.6 times more often than tests from LabCorp or Quest (P<0.0001). The percentages for measurements outside their normal range were 8.3% for LabCorp, 7.5% for Quest, and 12.2% for Theranos.
In addition, the researchers noted that, although they controlled subjects’ eating and physical activity, data from blood samples collected earlier in the day were sometimes significantly different from samples taken from the same subjects later in the day.
There were significant difference between measurements collected at time points 1 and 2 vs time points 3 and 4 for 13 of the 22 tests (P<0.002).
“These testing disparities occurred despite rigorous laboratory certification and proficiency standards designed to ensure consistency,” said study author Eric Schadt, PhD, of Mount Sinai.
“Our results suggest the need for greater transparency in lab technologies and procedures, as well as a much more thorough investigation of biological mechanisms that may contribute to more dynamic levels than we currently understand.” ![]()
iPSCs can differentiate into functional lymphocytes

Image from the Salk Institute
Researchers say they have generated induced pluripotent stem cells (iPSCs) that can differentiate into multiple lineages of functional lymphocytes.
The team noted that lymphohematopoietic stem cells (L-HSCs) generated from self-somatic cell-derived iPSCs could potentially be used to treat hematologic disorders, but no one has generated “truly functional” L-HSCs from iPSCs.
So the researchers set out to determine whether iPSCs have the inherent potential to generate multiple lineages of functional, terminally differentiated lymphocytes.
They described this work in Stem Cells and Development.
The researchers said they used tetraploid embryo complementation to provide a normal environment for the differentiation of L-HSCs from iPSCs and embryonic stem cells (ESCs). The team then compared lymphocytes derived from iPSCs, ESCs, and naïve isogenic C57BL/6 mice.
The researchers found that iPSC-derived lymphocytes expressed normal levels of major histocompatibility complex-I. Levels were comparable in iPSC-derived lymphocytes, ESC-derived lymphocytes, and lymphocytes from the control mice.
In addition, iPSC-derived lymphocytes were able to differentiate into multiple cell types—CD4+ T cells, CD8+ T cells, regulatory T cells, B cells, and natural killer cells.
Lymphocytes generated from iPSCs and lymphocytes generated from ESCs had the same capacity as lymphocytes from the control mice to proliferate and secrete chemical signals, such as cytokines.
All 3 types of lymphocytes proliferated under allogeneic stimulation but not under syngeneic stimulation. And the researchers found similar levels of IL-2, IL-4, IL-6, IL-10, IL-17, TNF, and IFN-γ in iPSC, ESC, and C57BL/6 lymphocyte culture supernatants.
The team also found that lymphocytes generated by iPSC-derived bone marrow cells could repopulate the hematopoietic systems of lethally irradiated recipient mice.
The iPSC bone marrow cells proved as effective as ESC-derived bone marrow cells and wild-type bone marrow cells. All 3 types of cells negated lymphocyte storage exhaustion in the spleen and peripheral blood.
In addition, there were no major phenotypic or behavioral abnormalities in any of the mice more than 1 month after cell transplantation.
The researchers said this work shows that truly functional lymphocytes can be generated from iPSCs, and it supports the clinical application of iPSC technology to develop treatments for hematologic disorders. ![]()

Image from the Salk Institute
Researchers say they have generated induced pluripotent stem cells (iPSCs) that can differentiate into multiple lineages of functional lymphocytes.
The team noted that lymphohematopoietic stem cells (L-HSCs) generated from self-somatic cell-derived iPSCs could potentially be used to treat hematologic disorders, but no one has generated “truly functional” L-HSCs from iPSCs.
So the researchers set out to determine whether iPSCs have the inherent potential to generate multiple lineages of functional, terminally differentiated lymphocytes.
They described this work in Stem Cells and Development.
The researchers said they used tetraploid embryo complementation to provide a normal environment for the differentiation of L-HSCs from iPSCs and embryonic stem cells (ESCs). The team then compared lymphocytes derived from iPSCs, ESCs, and naïve isogenic C57BL/6 mice.
The researchers found that iPSC-derived lymphocytes expressed normal levels of major histocompatibility complex-I. Levels were comparable in iPSC-derived lymphocytes, ESC-derived lymphocytes, and lymphocytes from the control mice.
In addition, iPSC-derived lymphocytes were able to differentiate into multiple cell types—CD4+ T cells, CD8+ T cells, regulatory T cells, B cells, and natural killer cells.
Lymphocytes generated from iPSCs and lymphocytes generated from ESCs had the same capacity as lymphocytes from the control mice to proliferate and secrete chemical signals, such as cytokines.
All 3 types of lymphocytes proliferated under allogeneic stimulation but not under syngeneic stimulation. And the researchers found similar levels of IL-2, IL-4, IL-6, IL-10, IL-17, TNF, and IFN-γ in iPSC, ESC, and C57BL/6 lymphocyte culture supernatants.
The team also found that lymphocytes generated by iPSC-derived bone marrow cells could repopulate the hematopoietic systems of lethally irradiated recipient mice.
The iPSC bone marrow cells proved as effective as ESC-derived bone marrow cells and wild-type bone marrow cells. All 3 types of cells negated lymphocyte storage exhaustion in the spleen and peripheral blood.
In addition, there were no major phenotypic or behavioral abnormalities in any of the mice more than 1 month after cell transplantation.
The researchers said this work shows that truly functional lymphocytes can be generated from iPSCs, and it supports the clinical application of iPSC technology to develop treatments for hematologic disorders. ![]()

Image from the Salk Institute
Researchers say they have generated induced pluripotent stem cells (iPSCs) that can differentiate into multiple lineages of functional lymphocytes.
The team noted that lymphohematopoietic stem cells (L-HSCs) generated from self-somatic cell-derived iPSCs could potentially be used to treat hematologic disorders, but no one has generated “truly functional” L-HSCs from iPSCs.
So the researchers set out to determine whether iPSCs have the inherent potential to generate multiple lineages of functional, terminally differentiated lymphocytes.
They described this work in Stem Cells and Development.
The researchers said they used tetraploid embryo complementation to provide a normal environment for the differentiation of L-HSCs from iPSCs and embryonic stem cells (ESCs). The team then compared lymphocytes derived from iPSCs, ESCs, and naïve isogenic C57BL/6 mice.
The researchers found that iPSC-derived lymphocytes expressed normal levels of major histocompatibility complex-I. Levels were comparable in iPSC-derived lymphocytes, ESC-derived lymphocytes, and lymphocytes from the control mice.
In addition, iPSC-derived lymphocytes were able to differentiate into multiple cell types—CD4+ T cells, CD8+ T cells, regulatory T cells, B cells, and natural killer cells.
Lymphocytes generated from iPSCs and lymphocytes generated from ESCs had the same capacity as lymphocytes from the control mice to proliferate and secrete chemical signals, such as cytokines.
All 3 types of lymphocytes proliferated under allogeneic stimulation but not under syngeneic stimulation. And the researchers found similar levels of IL-2, IL-4, IL-6, IL-10, IL-17, TNF, and IFN-γ in iPSC, ESC, and C57BL/6 lymphocyte culture supernatants.
The team also found that lymphocytes generated by iPSC-derived bone marrow cells could repopulate the hematopoietic systems of lethally irradiated recipient mice.
The iPSC bone marrow cells proved as effective as ESC-derived bone marrow cells and wild-type bone marrow cells. All 3 types of cells negated lymphocyte storage exhaustion in the spleen and peripheral blood.
In addition, there were no major phenotypic or behavioral abnormalities in any of the mice more than 1 month after cell transplantation.
The researchers said this work shows that truly functional lymphocytes can be generated from iPSCs, and it supports the clinical application of iPSC technology to develop treatments for hematologic disorders. ![]()
Research helps explain how malaria evolved

Photo by Holly Lutz
A study published in Molecular Phylogenetics and Evolution has revealed a new hypothesis on the evolution of malaria.
Researchers tested malarial DNA found in birds, bats, and other small mammals from 5 East African countries and found evidence to suggest that malaria has its roots in bird hosts.
It then spread to bats and on to other mammals.
“We can’t begin to understand how malaria spread to humans until we understand its evolutionary history,” said Holly Lutz, a doctoral candidate at Cornell University in Ithaca, New York.
“In learning about its past, we may be better able to understand the effects it has on us.”
Lutz and her colleagues took blood samples from hundreds of East African birds, bats, and other small mammals and screened the blood for malaria parasites.
When they found malaria, the team took samples of the parasites’ DNA and sequenced it to identify mutations in the genetic code. From there, the researchers performed phylogenetic analyses to determine how different malaria species are related.
In analyzing the genetic codes of the parasites, the team was able to find places where the DNA differed from one species to the next. Then, the researchers used computing software to determine how the different species evolved and how they’re related to each other.
“[B]y looking at patterns of mutations in the DNA of the different malaria species, we’re able to see when it branched off from one host group into another,” Lutz explained. “It started out as a parasite in birds, and then it evolved to colonize bats, and from there, it’s evolved to affect other mammals.”
In addition to shedding light on the way malaria was able to evolve and spread, the study provides information about the manner in which animals and their parasites are connected.
“Each of these individual vertebrates is an ecosystem in and of itself,” Lutz said. “In learning more about how parasites live within their hosts, who is infecting who, and how these organisms coexist in these living, breathing ecosystems, we can learn more about how they are connected to and affected by the natural environments that we share with animals and plants.”
The researchers noted that this study doesn’t have direct implications for malaria treatment in humans. However, the team believes that having a better understanding of malaria’s evolutionary history could help scientists anticipate how it will change and evolve in the future. ![]()

Photo by Holly Lutz
A study published in Molecular Phylogenetics and Evolution has revealed a new hypothesis on the evolution of malaria.
Researchers tested malarial DNA found in birds, bats, and other small mammals from 5 East African countries and found evidence to suggest that malaria has its roots in bird hosts.
It then spread to bats and on to other mammals.
“We can’t begin to understand how malaria spread to humans until we understand its evolutionary history,” said Holly Lutz, a doctoral candidate at Cornell University in Ithaca, New York.
“In learning about its past, we may be better able to understand the effects it has on us.”
Lutz and her colleagues took blood samples from hundreds of East African birds, bats, and other small mammals and screened the blood for malaria parasites.
When they found malaria, the team took samples of the parasites’ DNA and sequenced it to identify mutations in the genetic code. From there, the researchers performed phylogenetic analyses to determine how different malaria species are related.
In analyzing the genetic codes of the parasites, the team was able to find places where the DNA differed from one species to the next. Then, the researchers used computing software to determine how the different species evolved and how they’re related to each other.
“[B]y looking at patterns of mutations in the DNA of the different malaria species, we’re able to see when it branched off from one host group into another,” Lutz explained. “It started out as a parasite in birds, and then it evolved to colonize bats, and from there, it’s evolved to affect other mammals.”
In addition to shedding light on the way malaria was able to evolve and spread, the study provides information about the manner in which animals and their parasites are connected.
“Each of these individual vertebrates is an ecosystem in and of itself,” Lutz said. “In learning more about how parasites live within their hosts, who is infecting who, and how these organisms coexist in these living, breathing ecosystems, we can learn more about how they are connected to and affected by the natural environments that we share with animals and plants.”
The researchers noted that this study doesn’t have direct implications for malaria treatment in humans. However, the team believes that having a better understanding of malaria’s evolutionary history could help scientists anticipate how it will change and evolve in the future. ![]()

Photo by Holly Lutz
A study published in Molecular Phylogenetics and Evolution has revealed a new hypothesis on the evolution of malaria.
Researchers tested malarial DNA found in birds, bats, and other small mammals from 5 East African countries and found evidence to suggest that malaria has its roots in bird hosts.
It then spread to bats and on to other mammals.
“We can’t begin to understand how malaria spread to humans until we understand its evolutionary history,” said Holly Lutz, a doctoral candidate at Cornell University in Ithaca, New York.
“In learning about its past, we may be better able to understand the effects it has on us.”
Lutz and her colleagues took blood samples from hundreds of East African birds, bats, and other small mammals and screened the blood for malaria parasites.
When they found malaria, the team took samples of the parasites’ DNA and sequenced it to identify mutations in the genetic code. From there, the researchers performed phylogenetic analyses to determine how different malaria species are related.
In analyzing the genetic codes of the parasites, the team was able to find places where the DNA differed from one species to the next. Then, the researchers used computing software to determine how the different species evolved and how they’re related to each other.
“[B]y looking at patterns of mutations in the DNA of the different malaria species, we’re able to see when it branched off from one host group into another,” Lutz explained. “It started out as a parasite in birds, and then it evolved to colonize bats, and from there, it’s evolved to affect other mammals.”
In addition to shedding light on the way malaria was able to evolve and spread, the study provides information about the manner in which animals and their parasites are connected.
“Each of these individual vertebrates is an ecosystem in and of itself,” Lutz said. “In learning more about how parasites live within their hosts, who is infecting who, and how these organisms coexist in these living, breathing ecosystems, we can learn more about how they are connected to and affected by the natural environments that we share with animals and plants.”
The researchers noted that this study doesn’t have direct implications for malaria treatment in humans. However, the team believes that having a better understanding of malaria’s evolutionary history could help scientists anticipate how it will change and evolve in the future. ![]()
Team traces evolution of malaria

Photo courtesy of
Sesh Sundararaman,
University of Pennsylvania
By studying malaria parasites found in chimpanzees, researchers believe they have gained new insights regarding a malaria parasite that infects humans.
The team used a selective amplification technique to sequence the genomes of 2 divergent Plasmodium species, P reichenowi and P gaboni, from chimpanzee blood.
This revealed clues about the evolution and pathogenicity of P falciparum, the deadliest malaria parasite that affects humans.
The researchers described this work in Nature Communications.
They noted that African apes harbor at least 6 Plasmodium species that have been classified into a separate subgenus, called Laverania. Three of these Laverania species, including P reichenowi and P gaboni, reside in chimps.
Three others—including P praefalciparum, which gave rise to P falciparum—reside in gorillas. The gorilla origin of P falciparum was discovered several years ago by this same group of investigators.
“We want to know why Plasmodium falciparum is so deadly,” said Beatrice Hahn, MD, of the University of Pennsylvania in Philadelphia.
“The answer must lie in the blueprint—the genome—of its chimpanzee and gorilla cousins. We also want to know how and when the gorilla precursor of Plasmodium falciparum jumped into humans and why this happened only once.”
In an attempt to answer these questions, Dr Hahn and her colleagues used their selective amplification method to sequence Laverania genomes.
They used small amounts of unprocessed blood collected during routine health screens of chimpanzees living in sanctuaries. With their technique, the team found they could generate “high-quality” Laverania genome sequences.
The researchers said the chimpanzee parasite genomes contain information about the evolutionary origins of the malaria parasites infecting humans. One of the first things to emerge from genome-wide analyses was that the parasites represent distinct, non-interbreeding species.
In addition, members of each chimpanzee parasite species display about 10 times more genetic diversity than human parasites.
“The chimpanzee parasites really highlight the lack of diversity in Plasmodium falciparum,” said Paul Sharp, PhD, of the University of Edinburgh in the UK.
“This is most likely because these parasites went through a severe bottleneck when first transmitted to humans, perhaps within the past 10,000 years.”
By comparing the different parasite genomes, the researchers found an expansion of a multi-gene family, which governs red blood cell remodeling and therefore helps the parasite to evade host immune cells and clearance by the spleen.
“The remodeling process is a key part of severe malaria pathology in human Plasmodium falciparum infections,” said Julian Rayner, PhD, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“The expansion of this gene family from a single gene in all other Plasmodium parasites to up to 21 genes in Laverania suggests that remodeling evolved early in the radiation of this group of primate parasites and contributed not only to their unique biology but perhaps also to their successful expansion.”
“We also found a short region of the genome, including 2 essential invasion genes, where Plasmodium falciparum was much more different from its close relatives than we expected,” said Lindsey Plenderleith, PhD, of the University of Edinburgh.
Further analysis yielded the surprising finding that this fragment of DNA was horizontally transferred—from one species to another—into the gorilla ancestor of P falciparum.
“It is tempting to speculate that this unusual event somehow predisposed the precursor of Plasmodium falciparum to colonize humans,” Dr Hahn said. “However, this gene transfer clearly is not the entire story.”
Although the origin of P falciparum is considered well-established, nothing is known about the circumstances that led to its emergence.
“Coaxing entire parasite genome sequences out of small quantities of unprocessed ape blood will help us to better understand what happened and whether it can happen again,” said Sesh Sundararaman, an MD/PhD student at the University of Pennsylvania.
The team plans, as a next step, to use their select genome amplification technique to sequence additional ape parasite genomes to identify host-specific interactions and transmission requirements. They believe this would reveal vulnerabilities that might be exploited to combat malaria in humans. ![]()

Photo courtesy of
Sesh Sundararaman,
University of Pennsylvania
By studying malaria parasites found in chimpanzees, researchers believe they have gained new insights regarding a malaria parasite that infects humans.
The team used a selective amplification technique to sequence the genomes of 2 divergent Plasmodium species, P reichenowi and P gaboni, from chimpanzee blood.
This revealed clues about the evolution and pathogenicity of P falciparum, the deadliest malaria parasite that affects humans.
The researchers described this work in Nature Communications.
They noted that African apes harbor at least 6 Plasmodium species that have been classified into a separate subgenus, called Laverania. Three of these Laverania species, including P reichenowi and P gaboni, reside in chimps.
Three others—including P praefalciparum, which gave rise to P falciparum—reside in gorillas. The gorilla origin of P falciparum was discovered several years ago by this same group of investigators.
“We want to know why Plasmodium falciparum is so deadly,” said Beatrice Hahn, MD, of the University of Pennsylvania in Philadelphia.
“The answer must lie in the blueprint—the genome—of its chimpanzee and gorilla cousins. We also want to know how and when the gorilla precursor of Plasmodium falciparum jumped into humans and why this happened only once.”
In an attempt to answer these questions, Dr Hahn and her colleagues used their selective amplification method to sequence Laverania genomes.
They used small amounts of unprocessed blood collected during routine health screens of chimpanzees living in sanctuaries. With their technique, the team found they could generate “high-quality” Laverania genome sequences.
The researchers said the chimpanzee parasite genomes contain information about the evolutionary origins of the malaria parasites infecting humans. One of the first things to emerge from genome-wide analyses was that the parasites represent distinct, non-interbreeding species.
In addition, members of each chimpanzee parasite species display about 10 times more genetic diversity than human parasites.
“The chimpanzee parasites really highlight the lack of diversity in Plasmodium falciparum,” said Paul Sharp, PhD, of the University of Edinburgh in the UK.
“This is most likely because these parasites went through a severe bottleneck when first transmitted to humans, perhaps within the past 10,000 years.”
By comparing the different parasite genomes, the researchers found an expansion of a multi-gene family, which governs red blood cell remodeling and therefore helps the parasite to evade host immune cells and clearance by the spleen.
“The remodeling process is a key part of severe malaria pathology in human Plasmodium falciparum infections,” said Julian Rayner, PhD, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“The expansion of this gene family from a single gene in all other Plasmodium parasites to up to 21 genes in Laverania suggests that remodeling evolved early in the radiation of this group of primate parasites and contributed not only to their unique biology but perhaps also to their successful expansion.”
“We also found a short region of the genome, including 2 essential invasion genes, where Plasmodium falciparum was much more different from its close relatives than we expected,” said Lindsey Plenderleith, PhD, of the University of Edinburgh.
Further analysis yielded the surprising finding that this fragment of DNA was horizontally transferred—from one species to another—into the gorilla ancestor of P falciparum.
“It is tempting to speculate that this unusual event somehow predisposed the precursor of Plasmodium falciparum to colonize humans,” Dr Hahn said. “However, this gene transfer clearly is not the entire story.”
Although the origin of P falciparum is considered well-established, nothing is known about the circumstances that led to its emergence.
“Coaxing entire parasite genome sequences out of small quantities of unprocessed ape blood will help us to better understand what happened and whether it can happen again,” said Sesh Sundararaman, an MD/PhD student at the University of Pennsylvania.
The team plans, as a next step, to use their select genome amplification technique to sequence additional ape parasite genomes to identify host-specific interactions and transmission requirements. They believe this would reveal vulnerabilities that might be exploited to combat malaria in humans. ![]()

Photo courtesy of
Sesh Sundararaman,
University of Pennsylvania
By studying malaria parasites found in chimpanzees, researchers believe they have gained new insights regarding a malaria parasite that infects humans.
The team used a selective amplification technique to sequence the genomes of 2 divergent Plasmodium species, P reichenowi and P gaboni, from chimpanzee blood.
This revealed clues about the evolution and pathogenicity of P falciparum, the deadliest malaria parasite that affects humans.
The researchers described this work in Nature Communications.
They noted that African apes harbor at least 6 Plasmodium species that have been classified into a separate subgenus, called Laverania. Three of these Laverania species, including P reichenowi and P gaboni, reside in chimps.
Three others—including P praefalciparum, which gave rise to P falciparum—reside in gorillas. The gorilla origin of P falciparum was discovered several years ago by this same group of investigators.
“We want to know why Plasmodium falciparum is so deadly,” said Beatrice Hahn, MD, of the University of Pennsylvania in Philadelphia.
“The answer must lie in the blueprint—the genome—of its chimpanzee and gorilla cousins. We also want to know how and when the gorilla precursor of Plasmodium falciparum jumped into humans and why this happened only once.”
In an attempt to answer these questions, Dr Hahn and her colleagues used their selective amplification method to sequence Laverania genomes.
They used small amounts of unprocessed blood collected during routine health screens of chimpanzees living in sanctuaries. With their technique, the team found they could generate “high-quality” Laverania genome sequences.
The researchers said the chimpanzee parasite genomes contain information about the evolutionary origins of the malaria parasites infecting humans. One of the first things to emerge from genome-wide analyses was that the parasites represent distinct, non-interbreeding species.
In addition, members of each chimpanzee parasite species display about 10 times more genetic diversity than human parasites.
“The chimpanzee parasites really highlight the lack of diversity in Plasmodium falciparum,” said Paul Sharp, PhD, of the University of Edinburgh in the UK.
“This is most likely because these parasites went through a severe bottleneck when first transmitted to humans, perhaps within the past 10,000 years.”
By comparing the different parasite genomes, the researchers found an expansion of a multi-gene family, which governs red blood cell remodeling and therefore helps the parasite to evade host immune cells and clearance by the spleen.
“The remodeling process is a key part of severe malaria pathology in human Plasmodium falciparum infections,” said Julian Rayner, PhD, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“The expansion of this gene family from a single gene in all other Plasmodium parasites to up to 21 genes in Laverania suggests that remodeling evolved early in the radiation of this group of primate parasites and contributed not only to their unique biology but perhaps also to their successful expansion.”
“We also found a short region of the genome, including 2 essential invasion genes, where Plasmodium falciparum was much more different from its close relatives than we expected,” said Lindsey Plenderleith, PhD, of the University of Edinburgh.
Further analysis yielded the surprising finding that this fragment of DNA was horizontally transferred—from one species to another—into the gorilla ancestor of P falciparum.
“It is tempting to speculate that this unusual event somehow predisposed the precursor of Plasmodium falciparum to colonize humans,” Dr Hahn said. “However, this gene transfer clearly is not the entire story.”
Although the origin of P falciparum is considered well-established, nothing is known about the circumstances that led to its emergence.
“Coaxing entire parasite genome sequences out of small quantities of unprocessed ape blood will help us to better understand what happened and whether it can happen again,” said Sesh Sundararaman, an MD/PhD student at the University of Pennsylvania.
The team plans, as a next step, to use their select genome amplification technique to sequence additional ape parasite genomes to identify host-specific interactions and transmission requirements. They believe this would reveal vulnerabilities that might be exploited to combat malaria in humans. ![]()
Parasite competition influences drug resistance
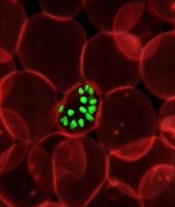
infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
Researchers say they have documented how competition among different malaria parasite strains in human hosts could influence the spread of drug resistance.
“We found that when hosts are co-infected with drug-resistant and drug-sensitive strains, both strains are competitively suppressed,” said Mary Bushman, of Emory University in Atlanta, Georgia.
“Antimalarial therapy, by clearing drug-sensitive parasites from mixed infections, may result in competitive release of resistant strains.”
Bushman and her colleagues described these findings in Proceedings of the Royal Society B.
The researchers focused on the parasite Plasmodium falciparum, which has developed resistance to former first-line therapies chloroquine and sulfadoxine-pyrimethamine.
“We’re now down to our last treatment, artemisinin combination therapy, or ACT, and resistance to that recently emerged in Southeast Asia,” Bushman said. “If ACT resistance continues to follow the same pattern, the world may soon be without reliable antimalarial drugs.”
In addition, people infected with P falciparum often have multiple strains of the parasite, especially in high-transmission areas such as sub-Saharan Africa, where infectious mosquito bites occur frequently. Many people have developed partial immunity, making asymptomatic infections common and further complicating control efforts.
With previous work in lab mice, the researchers found that competition between mixed strains of malaria parasites were a crucial determinant to the spread of resistance.
“In the mouse studies, we found that drug-sensitive parasites suppress resistant parasites,” said Jaap de Roode, PhD, of Emory University.
“We also found that by clearing these sensitive parasites with drugs, the resistant parasites had a big advantage, growing up to high numbers and transmitting to mosquitoes at high rates. Ever since doing that work, I have wanted to see if the same could apply to humans.”
To find out, Dr de Roode and his colleagues analyzed 1341 blood samples from untreated children with malaria living in Angola, Ghana, and Tanzania.
The researchers extracted the DNA of malaria parasites from the blood samples and used polymerase chain reaction technology to determine the densities of drug-resistant strains and drug-sensitive ones. About 15% of the samples had mixtures of both types.
Analyses showed that, in mixed-strain infections, densities of chloroquine-sensitive and chloroquine-resistant strains were reduced in the presence of competitors.
The results also showed that, in the absence of chloroquine, the resistant strains had lower densities than sensitive strains.
“The results were really clear cut, which rarely happens in human studies,” Bushman said. “We found almost complete consistency between the 3 data sets [divided by country].”
Bushman added that the tendency is to use a “one-size-fits-all” strategy for controlling malaria, but this research suggests that more tailored approaches are needed.
For example, a strategy of mass drug administration might be effective in a place with a low prevalence of malaria and less likelihood of mixed-strain infections. However, that same strategy might actually boost drug resistance without reducing the burden of disease in areas where most of the population is infected with multiple strains of malaria parasites.
“The epidemiology of malaria infection is different for different places and different conditions,” Bushman said. “We hope that our work will spur development of new strategies to minimize resistance while maximizing the benefits of control measures.”
However, more questions must be answered to guide the development of these new strategies.
“As a first step, we need to determine if the observed suppression of resistance in humans also results in reduced transmission to mosquitoes,” Dr de Roode said.
Another avenue to explore is resistance among patients who have received antimalarial treatment.
“We need to find out if drug treatment of people infected with malaria removes competition and gives resistance a boost, as we have found in mice before,” Dr de Roode noted. ![]()

infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
Researchers say they have documented how competition among different malaria parasite strains in human hosts could influence the spread of drug resistance.
“We found that when hosts are co-infected with drug-resistant and drug-sensitive strains, both strains are competitively suppressed,” said Mary Bushman, of Emory University in Atlanta, Georgia.
“Antimalarial therapy, by clearing drug-sensitive parasites from mixed infections, may result in competitive release of resistant strains.”
Bushman and her colleagues described these findings in Proceedings of the Royal Society B.
The researchers focused on the parasite Plasmodium falciparum, which has developed resistance to former first-line therapies chloroquine and sulfadoxine-pyrimethamine.
“We’re now down to our last treatment, artemisinin combination therapy, or ACT, and resistance to that recently emerged in Southeast Asia,” Bushman said. “If ACT resistance continues to follow the same pattern, the world may soon be without reliable antimalarial drugs.”
In addition, people infected with P falciparum often have multiple strains of the parasite, especially in high-transmission areas such as sub-Saharan Africa, where infectious mosquito bites occur frequently. Many people have developed partial immunity, making asymptomatic infections common and further complicating control efforts.
With previous work in lab mice, the researchers found that competition between mixed strains of malaria parasites were a crucial determinant to the spread of resistance.
“In the mouse studies, we found that drug-sensitive parasites suppress resistant parasites,” said Jaap de Roode, PhD, of Emory University.
“We also found that by clearing these sensitive parasites with drugs, the resistant parasites had a big advantage, growing up to high numbers and transmitting to mosquitoes at high rates. Ever since doing that work, I have wanted to see if the same could apply to humans.”
To find out, Dr de Roode and his colleagues analyzed 1341 blood samples from untreated children with malaria living in Angola, Ghana, and Tanzania.
The researchers extracted the DNA of malaria parasites from the blood samples and used polymerase chain reaction technology to determine the densities of drug-resistant strains and drug-sensitive ones. About 15% of the samples had mixtures of both types.
Analyses showed that, in mixed-strain infections, densities of chloroquine-sensitive and chloroquine-resistant strains were reduced in the presence of competitors.
The results also showed that, in the absence of chloroquine, the resistant strains had lower densities than sensitive strains.
“The results were really clear cut, which rarely happens in human studies,” Bushman said. “We found almost complete consistency between the 3 data sets [divided by country].”
Bushman added that the tendency is to use a “one-size-fits-all” strategy for controlling malaria, but this research suggests that more tailored approaches are needed.
For example, a strategy of mass drug administration might be effective in a place with a low prevalence of malaria and less likelihood of mixed-strain infections. However, that same strategy might actually boost drug resistance without reducing the burden of disease in areas where most of the population is infected with multiple strains of malaria parasites.
“The epidemiology of malaria infection is different for different places and different conditions,” Bushman said. “We hope that our work will spur development of new strategies to minimize resistance while maximizing the benefits of control measures.”
However, more questions must be answered to guide the development of these new strategies.
“As a first step, we need to determine if the observed suppression of resistance in humans also results in reduced transmission to mosquitoes,” Dr de Roode said.
Another avenue to explore is resistance among patients who have received antimalarial treatment.
“We need to find out if drug treatment of people infected with malaria removes competition and gives resistance a boost, as we have found in mice before,” Dr de Roode noted. ![]()

infecting a red blood cell
Image courtesy of St. Jude
Children’s Research Hospital
Researchers say they have documented how competition among different malaria parasite strains in human hosts could influence the spread of drug resistance.
“We found that when hosts are co-infected with drug-resistant and drug-sensitive strains, both strains are competitively suppressed,” said Mary Bushman, of Emory University in Atlanta, Georgia.
“Antimalarial therapy, by clearing drug-sensitive parasites from mixed infections, may result in competitive release of resistant strains.”
Bushman and her colleagues described these findings in Proceedings of the Royal Society B.
The researchers focused on the parasite Plasmodium falciparum, which has developed resistance to former first-line therapies chloroquine and sulfadoxine-pyrimethamine.
“We’re now down to our last treatment, artemisinin combination therapy, or ACT, and resistance to that recently emerged in Southeast Asia,” Bushman said. “If ACT resistance continues to follow the same pattern, the world may soon be without reliable antimalarial drugs.”
In addition, people infected with P falciparum often have multiple strains of the parasite, especially in high-transmission areas such as sub-Saharan Africa, where infectious mosquito bites occur frequently. Many people have developed partial immunity, making asymptomatic infections common and further complicating control efforts.
With previous work in lab mice, the researchers found that competition between mixed strains of malaria parasites were a crucial determinant to the spread of resistance.
“In the mouse studies, we found that drug-sensitive parasites suppress resistant parasites,” said Jaap de Roode, PhD, of Emory University.
“We also found that by clearing these sensitive parasites with drugs, the resistant parasites had a big advantage, growing up to high numbers and transmitting to mosquitoes at high rates. Ever since doing that work, I have wanted to see if the same could apply to humans.”
To find out, Dr de Roode and his colleagues analyzed 1341 blood samples from untreated children with malaria living in Angola, Ghana, and Tanzania.
The researchers extracted the DNA of malaria parasites from the blood samples and used polymerase chain reaction technology to determine the densities of drug-resistant strains and drug-sensitive ones. About 15% of the samples had mixtures of both types.
Analyses showed that, in mixed-strain infections, densities of chloroquine-sensitive and chloroquine-resistant strains were reduced in the presence of competitors.
The results also showed that, in the absence of chloroquine, the resistant strains had lower densities than sensitive strains.
“The results were really clear cut, which rarely happens in human studies,” Bushman said. “We found almost complete consistency between the 3 data sets [divided by country].”
Bushman added that the tendency is to use a “one-size-fits-all” strategy for controlling malaria, but this research suggests that more tailored approaches are needed.
For example, a strategy of mass drug administration might be effective in a place with a low prevalence of malaria and less likelihood of mixed-strain infections. However, that same strategy might actually boost drug resistance without reducing the burden of disease in areas where most of the population is infected with multiple strains of malaria parasites.
“The epidemiology of malaria infection is different for different places and different conditions,” Bushman said. “We hope that our work will spur development of new strategies to minimize resistance while maximizing the benefits of control measures.”
However, more questions must be answered to guide the development of these new strategies.
“As a first step, we need to determine if the observed suppression of resistance in humans also results in reduced transmission to mosquitoes,” Dr de Roode said.
Another avenue to explore is resistance among patients who have received antimalarial treatment.
“We need to find out if drug treatment of people infected with malaria removes competition and gives resistance a boost, as we have found in mice before,” Dr de Roode noted. ![]()
Use of dangerous drug combos on the rise

Photo by Rhoda Baer
A new study suggests that older adults in the US may be using potentially deadly combinations of prescription drugs, over-the-counter medications, and dietary supplements.
Investigators observed a significant increase in the proportion of adults ages 62 to 85 who were at risk of major drug-drug interactions—from 8% in 2005-2006 to 15% in 2010-2011.
Most of the combinations consisted of preventative cardiovascular medications such as antiplatelet drugs, statins, and omega-3 fish oil supplements.
Dima Mazen Qato, PharmD, of the University of Illinois at Chicago, and her colleagues reported these results in JAMA Internal Medicine.
The investigators examined changes in medication use in a nationally representative sample of adults between the ages of 62 and 85. The team conducted in-home interviews to accurately identify medications. They interviewed 2351 subjects in 2005-2006 and 2206 subjects in 2010-2011.
The results showed a significant increase in the proportion of subjects taking at least 1 prescription medication—from 84.1% in 2005-2006 to 87.7% in 2010-2011 (P=0.003).
The proportion of subjects taking at least 5 prescription medications rose significantly as well—from 30.6% to 35.8% (P=0.02).
The investigators said factors that may account for these increases include the implementation of Medicare Part D, changes in treatment guidelines, and the increased availability of generics for many commonly used drugs.
As an example, use of the statin simvastatin (Zocor)—which became available as a generic in 2006—rose from 10.3% in 2005-2006 to 22.5% in 2010-2011.
The use of over-the-counter medications declined from 44.4% in 2005-2006 to 37.9% in 2010-2011 (P<0.001), while the use of dietary supplements increased from 51.8% to 63.7% (P<0.001).
Drug-drug interactions
The investigators identified 15 potentially life-threatening combinations of the most commonly used medications and supplements in the study.
They found that 15.1% of subjects were at risk for a potential major drug-drug interaction in 2010-2011, compared with an estimated 8.4% in 2005-2006 (P<0.001).
Dr Qato noted that more than half of the potential interactions involved a nonprescription medication or dietary supplement.
Preventative cardiovascular medications such as statins (particularly, simvastatin), antiplatelet drugs (such as clopidogrel and aspirin), and supplements (specifically, omega-3 fish oil) accounted for the vast majority of these combinations.
“Many older patients seeking to improve their cardiovascular health are also regularly using interacting drug combinations that may worsen cardiovascular risk,” Dr Qato said.
“For example, the use of clopidogrel in combination with the proton-pump inhibitor omeprazole, aspirin, or naproxen—all over-the-counter medications—is associated with an increased risk of heart attacks, bleeding complications, or death. However, about 1.8%—or 1 million—older adults regularly use clopidogrel in interacting combinations.”
Dr Qato added that healthcare professionals should consider the adverse effects of commonly used prescription and nonprescription medication combinations when treating older adults and counsel patients about the risks.
“Improving safety in the use of interacting medication combinations has the potential to reduce preventable, potentially fatal, adverse drug events,” she said. ![]()

Photo by Rhoda Baer
A new study suggests that older adults in the US may be using potentially deadly combinations of prescription drugs, over-the-counter medications, and dietary supplements.
Investigators observed a significant increase in the proportion of adults ages 62 to 85 who were at risk of major drug-drug interactions—from 8% in 2005-2006 to 15% in 2010-2011.
Most of the combinations consisted of preventative cardiovascular medications such as antiplatelet drugs, statins, and omega-3 fish oil supplements.
Dima Mazen Qato, PharmD, of the University of Illinois at Chicago, and her colleagues reported these results in JAMA Internal Medicine.
The investigators examined changes in medication use in a nationally representative sample of adults between the ages of 62 and 85. The team conducted in-home interviews to accurately identify medications. They interviewed 2351 subjects in 2005-2006 and 2206 subjects in 2010-2011.
The results showed a significant increase in the proportion of subjects taking at least 1 prescription medication—from 84.1% in 2005-2006 to 87.7% in 2010-2011 (P=0.003).
The proportion of subjects taking at least 5 prescription medications rose significantly as well—from 30.6% to 35.8% (P=0.02).
The investigators said factors that may account for these increases include the implementation of Medicare Part D, changes in treatment guidelines, and the increased availability of generics for many commonly used drugs.
As an example, use of the statin simvastatin (Zocor)—which became available as a generic in 2006—rose from 10.3% in 2005-2006 to 22.5% in 2010-2011.
The use of over-the-counter medications declined from 44.4% in 2005-2006 to 37.9% in 2010-2011 (P<0.001), while the use of dietary supplements increased from 51.8% to 63.7% (P<0.001).
Drug-drug interactions
The investigators identified 15 potentially life-threatening combinations of the most commonly used medications and supplements in the study.
They found that 15.1% of subjects were at risk for a potential major drug-drug interaction in 2010-2011, compared with an estimated 8.4% in 2005-2006 (P<0.001).
Dr Qato noted that more than half of the potential interactions involved a nonprescription medication or dietary supplement.
Preventative cardiovascular medications such as statins (particularly, simvastatin), antiplatelet drugs (such as clopidogrel and aspirin), and supplements (specifically, omega-3 fish oil) accounted for the vast majority of these combinations.
“Many older patients seeking to improve their cardiovascular health are also regularly using interacting drug combinations that may worsen cardiovascular risk,” Dr Qato said.
“For example, the use of clopidogrel in combination with the proton-pump inhibitor omeprazole, aspirin, or naproxen—all over-the-counter medications—is associated with an increased risk of heart attacks, bleeding complications, or death. However, about 1.8%—or 1 million—older adults regularly use clopidogrel in interacting combinations.”
Dr Qato added that healthcare professionals should consider the adverse effects of commonly used prescription and nonprescription medication combinations when treating older adults and counsel patients about the risks.
“Improving safety in the use of interacting medication combinations has the potential to reduce preventable, potentially fatal, adverse drug events,” she said. ![]()

Photo by Rhoda Baer
A new study suggests that older adults in the US may be using potentially deadly combinations of prescription drugs, over-the-counter medications, and dietary supplements.
Investigators observed a significant increase in the proportion of adults ages 62 to 85 who were at risk of major drug-drug interactions—from 8% in 2005-2006 to 15% in 2010-2011.
Most of the combinations consisted of preventative cardiovascular medications such as antiplatelet drugs, statins, and omega-3 fish oil supplements.
Dima Mazen Qato, PharmD, of the University of Illinois at Chicago, and her colleagues reported these results in JAMA Internal Medicine.
The investigators examined changes in medication use in a nationally representative sample of adults between the ages of 62 and 85. The team conducted in-home interviews to accurately identify medications. They interviewed 2351 subjects in 2005-2006 and 2206 subjects in 2010-2011.
The results showed a significant increase in the proportion of subjects taking at least 1 prescription medication—from 84.1% in 2005-2006 to 87.7% in 2010-2011 (P=0.003).
The proportion of subjects taking at least 5 prescription medications rose significantly as well—from 30.6% to 35.8% (P=0.02).
The investigators said factors that may account for these increases include the implementation of Medicare Part D, changes in treatment guidelines, and the increased availability of generics for many commonly used drugs.
As an example, use of the statin simvastatin (Zocor)—which became available as a generic in 2006—rose from 10.3% in 2005-2006 to 22.5% in 2010-2011.
The use of over-the-counter medications declined from 44.4% in 2005-2006 to 37.9% in 2010-2011 (P<0.001), while the use of dietary supplements increased from 51.8% to 63.7% (P<0.001).
Drug-drug interactions
The investigators identified 15 potentially life-threatening combinations of the most commonly used medications and supplements in the study.
They found that 15.1% of subjects were at risk for a potential major drug-drug interaction in 2010-2011, compared with an estimated 8.4% in 2005-2006 (P<0.001).
Dr Qato noted that more than half of the potential interactions involved a nonprescription medication or dietary supplement.
Preventative cardiovascular medications such as statins (particularly, simvastatin), antiplatelet drugs (such as clopidogrel and aspirin), and supplements (specifically, omega-3 fish oil) accounted for the vast majority of these combinations.
“Many older patients seeking to improve their cardiovascular health are also regularly using interacting drug combinations that may worsen cardiovascular risk,” Dr Qato said.
“For example, the use of clopidogrel in combination with the proton-pump inhibitor omeprazole, aspirin, or naproxen—all over-the-counter medications—is associated with an increased risk of heart attacks, bleeding complications, or death. However, about 1.8%—or 1 million—older adults regularly use clopidogrel in interacting combinations.”
Dr Qato added that healthcare professionals should consider the adverse effects of commonly used prescription and nonprescription medication combinations when treating older adults and counsel patients about the risks.
“Improving safety in the use of interacting medication combinations has the potential to reduce preventable, potentially fatal, adverse drug events,” she said.
FDA proposes ban on most powdered medical gloves
Photo by Chonion Antoine
The US Food and Drug Administration (FDA) is proposing a ban on most powdered gloves in the US.
The proposed ban applies to powdered surgeon’s gloves, powdered patient examination gloves, and absorbable powder for lubricating a surgeon’s glove.
The FDA said these gloves pose an unreasonable and substantial risk of illness or injury to healthcare providers, patients, and other individuals who are exposed to them, which cannot be corrected through new or updated labeling.
“This ban is about protecting patients and healthcare professionals from a danger they might not even be aware of,” said Jeffrey Shuren, MD, director of FDA’s Center for Devices and Radiological Health.
“We take bans very seriously and only take this action when we feel it’s necessary to protect the public health.”
The FDA said powdered gloves are dangerous for a variety of reasons. In particular, aerosolized glove powder on natural rubber latex gloves, but not on synthetic powdered gloves, can carry proteins that may cause respiratory allergic reactions.
Although powdered synthetic gloves do not present the risk of allergic reactions, these gloves are associated with an extensive list of potentially serious adverse events, including severe airway inflammation, wound inflammation, and post-surgical adhesions. These side effects have been attributed to the use of glove powder with all types of gloves.
As these risks cannot be corrected through new or updated labeling, the FDA is moving forward with the proposal to ban these products, which—if finalized—would ultimately remove them from the marketplace completely.
In making the determination that these products are dangerous and present an unreasonable and substantial risk, the FDA considered all available evidence, which included a thorough review of the scientific literature and comments received on a February 2011 Federal Register Notice.
The FDA also conducted an economic analysis that showed a powdered glove ban would not cause a glove shortage, and the economic impact of a ban would not be significant. The ban is also not likely to impact medical practice, because many non-powdered protective glove options are currently available.
The FDA has determined that the banning standard would not apply to powdered radiographic protection gloves. The agency is not aware of any powdered radiographic protection gloves that are currently on the market.
Non-powdered surgeon gloves and non-powdered patient examination gloves will not be included in the ban and will remain Class I medical devices. Therefore, the FDA is also proposing amendments to their classification regulations to clarify that they apply only to non-powdered gloves.
The proposed rule is available online at www.regulations.gov for public comment for 90 days.

Photo by Chonion Antoine
The US Food and Drug Administration (FDA) is proposing a ban on most powdered gloves in the US.
The proposed ban applies to powdered surgeon’s gloves, powdered patient examination gloves, and absorbable powder for lubricating a surgeon’s glove.
The FDA said these gloves pose an unreasonable and substantial risk of illness or injury to healthcare providers, patients, and other individuals who are exposed to them, which cannot be corrected through new or updated labeling.
“This ban is about protecting patients and healthcare professionals from a danger they might not even be aware of,” said Jeffrey Shuren, MD, director of FDA’s Center for Devices and Radiological Health.
“We take bans very seriously and only take this action when we feel it’s necessary to protect the public health.”
The FDA said powdered gloves are dangerous for a variety of reasons. In particular, aerosolized glove powder on natural rubber latex gloves, but not on synthetic powdered gloves, can carry proteins that may cause respiratory allergic reactions.
Although powdered synthetic gloves do not present the risk of allergic reactions, these gloves are associated with an extensive list of potentially serious adverse events, including severe airway inflammation, wound inflammation, and post-surgical adhesions. These side effects have been attributed to the use of glove powder with all types of gloves.
As these risks cannot be corrected through new or updated labeling, the FDA is moving forward with the proposal to ban these products, which—if finalized—would ultimately remove them from the marketplace completely.
In making the determination that these products are dangerous and present an unreasonable and substantial risk, the FDA considered all available evidence, which included a thorough review of the scientific literature and comments received on a February 2011 Federal Register Notice.
The FDA also conducted an economic analysis that showed a powdered glove ban would not cause a glove shortage, and the economic impact of a ban would not be significant. The ban is also not likely to impact medical practice, because many non-powdered protective glove options are currently available.
The FDA has determined that the banning standard would not apply to powdered radiographic protection gloves. The agency is not aware of any powdered radiographic protection gloves that are currently on the market.
Non-powdered surgeon gloves and non-powdered patient examination gloves will not be included in the ban and will remain Class I medical devices. Therefore, the FDA is also proposing amendments to their classification regulations to clarify that they apply only to non-powdered gloves.
The proposed rule is available online at www.regulations.gov for public comment for 90 days.

Photo by Chonion Antoine
The US Food and Drug Administration (FDA) is proposing a ban on most powdered gloves in the US.
The proposed ban applies to powdered surgeon’s gloves, powdered patient examination gloves, and absorbable powder for lubricating a surgeon’s glove.
The FDA said these gloves pose an unreasonable and substantial risk of illness or injury to healthcare providers, patients, and other individuals who are exposed to them, which cannot be corrected through new or updated labeling.
“This ban is about protecting patients and healthcare professionals from a danger they might not even be aware of,” said Jeffrey Shuren, MD, director of FDA’s Center for Devices and Radiological Health.
“We take bans very seriously and only take this action when we feel it’s necessary to protect the public health.”
The FDA said powdered gloves are dangerous for a variety of reasons. In particular, aerosolized glove powder on natural rubber latex gloves, but not on synthetic powdered gloves, can carry proteins that may cause respiratory allergic reactions.
Although powdered synthetic gloves do not present the risk of allergic reactions, these gloves are associated with an extensive list of potentially serious adverse events, including severe airway inflammation, wound inflammation, and post-surgical adhesions. These side effects have been attributed to the use of glove powder with all types of gloves.
As these risks cannot be corrected through new or updated labeling, the FDA is moving forward with the proposal to ban these products, which—if finalized—would ultimately remove them from the marketplace completely.
In making the determination that these products are dangerous and present an unreasonable and substantial risk, the FDA considered all available evidence, which included a thorough review of the scientific literature and comments received on a February 2011 Federal Register Notice.
The FDA also conducted an economic analysis that showed a powdered glove ban would not cause a glove shortage, and the economic impact of a ban would not be significant. The ban is also not likely to impact medical practice, because many non-powdered protective glove options are currently available.
The FDA has determined that the banning standard would not apply to powdered radiographic protection gloves. The agency is not aware of any powdered radiographic protection gloves that are currently on the market.
Non-powdered surgeon gloves and non-powdered patient examination gloves will not be included in the ban and will remain Class I medical devices. Therefore, the FDA is also proposing amendments to their classification regulations to clarify that they apply only to non-powdered gloves.
The proposed rule is available online at www.regulations.gov for public comment for 90 days.
FDA authorizes new test for Zika virus

Photo by Graham Colm
In response to a request from the US Centers for Disease Control and Prevention (CDC), the US Food and Drug Administration (FDA) has issued an Emergency Use Authorization (EUA) for the Trioplex Real-time RT-PCR Assay, a tool that can be used to detect Zika virus.
The assay allows doctors to tell if an individual is infected with chikungunya, dengue, or Zika virus using a single test, instead of having to perform 3 separate tests to identify the infection.
The Trioplex Real-time RT-PCR Assay can be used to detect virus RNA in serum, cerebrospinal fluid, urine, and amniotic fluid specimens.
The CDC hopes this EUA will allow the agency to more rapidly perform testing to detect acute Zika virus infection.
An EUA allows the use of unapproved medical products or unapproved uses of approved medical products in an emergency. The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The CDC said it will begin distributing the Trioplex Real-time RT-PCR Assay during the next 2 weeks to qualified laboratories in the Laboratory Response Network, an integrated network of domestic and international laboratories that respond to public health emergencies.
The test will not be available in US hospitals or other primary care settings.
Last month, the FDA issued an EUA for a different test used to detect the Zika virus, the Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA).
This test was distributed to qualified laboratories in the Laboratory Response Network but was not made available in US hospitals or other primary care settings.

Photo by Graham Colm
In response to a request from the US Centers for Disease Control and Prevention (CDC), the US Food and Drug Administration (FDA) has issued an Emergency Use Authorization (EUA) for the Trioplex Real-time RT-PCR Assay, a tool that can be used to detect Zika virus.
The assay allows doctors to tell if an individual is infected with chikungunya, dengue, or Zika virus using a single test, instead of having to perform 3 separate tests to identify the infection.
The Trioplex Real-time RT-PCR Assay can be used to detect virus RNA in serum, cerebrospinal fluid, urine, and amniotic fluid specimens.
The CDC hopes this EUA will allow the agency to more rapidly perform testing to detect acute Zika virus infection.
An EUA allows the use of unapproved medical products or unapproved uses of approved medical products in an emergency. The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The CDC said it will begin distributing the Trioplex Real-time RT-PCR Assay during the next 2 weeks to qualified laboratories in the Laboratory Response Network, an integrated network of domestic and international laboratories that respond to public health emergencies.
The test will not be available in US hospitals or other primary care settings.
Last month, the FDA issued an EUA for a different test used to detect the Zika virus, the Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA).
This test was distributed to qualified laboratories in the Laboratory Response Network but was not made available in US hospitals or other primary care settings.

Photo by Graham Colm
In response to a request from the US Centers for Disease Control and Prevention (CDC), the US Food and Drug Administration (FDA) has issued an Emergency Use Authorization (EUA) for the Trioplex Real-time RT-PCR Assay, a tool that can be used to detect Zika virus.
The assay allows doctors to tell if an individual is infected with chikungunya, dengue, or Zika virus using a single test, instead of having to perform 3 separate tests to identify the infection.
The Trioplex Real-time RT-PCR Assay can be used to detect virus RNA in serum, cerebrospinal fluid, urine, and amniotic fluid specimens.
The CDC hopes this EUA will allow the agency to more rapidly perform testing to detect acute Zika virus infection.
An EUA allows the use of unapproved medical products or unapproved uses of approved medical products in an emergency. The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
The CDC said it will begin distributing the Trioplex Real-time RT-PCR Assay during the next 2 weeks to qualified laboratories in the Laboratory Response Network, an integrated network of domestic and international laboratories that respond to public health emergencies.
The test will not be available in US hospitals or other primary care settings.
Last month, the FDA issued an EUA for a different test used to detect the Zika virus, the Zika IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA).
This test was distributed to qualified laboratories in the Laboratory Response Network but was not made available in US hospitals or other primary care settings.
Bloodstream infection linked to antinausea drug
Researchers say they have discovered the source of a bloodstream infection observed in more than 50 South American cancer patients.
Using whole-genome sequence typing (WGST), the team was able to link infection with the fungus Sarocladium kiliense to a tainted antinausea medication, ondansetron, that was given to cancer patients in Chile and Colombia.
This work is described in Emerging Infectious Diseases.
“Contamination of medical products, particularly with environmental fungi, poses growing concern and a public health threat, especially in vulnerable populations such as cancer patients,” said study author David Engelthaler, PhD, of The Translational Genomics Research Institute in Flagstaff, Arizona.
“Increased vigilance and the use of advanced technologies are needed to rapidly identify the likely sources of infection to efficiently guide epidemiologic investigations and initiate appropriate control measures.”
The S kiliense bloodstream-infection outbreak, which occurred from June 2013 through January 2014, included a cluster of cases at 8 hospitals in Santiago, Chile.
All of the patients received the same 4 intravenous medications. But only the antinausea medication ondansetron was given exclusively to cancer patients.
All of the patients infected with S kiliense received ondansetron from the same source, a pharmaceutical company called Vitrofarma SA (specifically, Plant No. 8 in Bogotá, Colombia). The drug was imported by LabVitales Chile SA and distributed by Pharma Isa Ltda.
Two of 3 lots of unopened ondansetron, tested by the Chilean Ministry of Health, yielded vials contaminated with S kiliense, forcing a recall of all ondansetron in Chile that was made by Vitrofarma SA.
Subsequently, Colombian officials discovered 14 other cases in which patients who were given ondansetron from Vitrofarma SA were infected with S kiliense.
S kiliense has been implicated in healthcare-related infections before, but the lack of available typing methods has precluded the ability to substantiate sources.
“The use of WGST to investigate fungal outbreaks has become integral to epidemiologic investigations,” Dr Engelthaler said. “Our WGST analysis demonstrated that the patient isolates from Chile and Colombia were nearly genetically indistinguishable from those recovered from the unopened medication vials, indicating the likely presence of a single-source infection.”

Researchers say they have discovered the source of a bloodstream infection observed in more than 50 South American cancer patients.
Using whole-genome sequence typing (WGST), the team was able to link infection with the fungus Sarocladium kiliense to a tainted antinausea medication, ondansetron, that was given to cancer patients in Chile and Colombia.
This work is described in Emerging Infectious Diseases.
“Contamination of medical products, particularly with environmental fungi, poses growing concern and a public health threat, especially in vulnerable populations such as cancer patients,” said study author David Engelthaler, PhD, of The Translational Genomics Research Institute in Flagstaff, Arizona.
“Increased vigilance and the use of advanced technologies are needed to rapidly identify the likely sources of infection to efficiently guide epidemiologic investigations and initiate appropriate control measures.”
The S kiliense bloodstream-infection outbreak, which occurred from June 2013 through January 2014, included a cluster of cases at 8 hospitals in Santiago, Chile.
All of the patients received the same 4 intravenous medications. But only the antinausea medication ondansetron was given exclusively to cancer patients.
All of the patients infected with S kiliense received ondansetron from the same source, a pharmaceutical company called Vitrofarma SA (specifically, Plant No. 8 in Bogotá, Colombia). The drug was imported by LabVitales Chile SA and distributed by Pharma Isa Ltda.
Two of 3 lots of unopened ondansetron, tested by the Chilean Ministry of Health, yielded vials contaminated with S kiliense, forcing a recall of all ondansetron in Chile that was made by Vitrofarma SA.
Subsequently, Colombian officials discovered 14 other cases in which patients who were given ondansetron from Vitrofarma SA were infected with S kiliense.
S kiliense has been implicated in healthcare-related infections before, but the lack of available typing methods has precluded the ability to substantiate sources.
“The use of WGST to investigate fungal outbreaks has become integral to epidemiologic investigations,” Dr Engelthaler said. “Our WGST analysis demonstrated that the patient isolates from Chile and Colombia were nearly genetically indistinguishable from those recovered from the unopened medication vials, indicating the likely presence of a single-source infection.”

Researchers say they have discovered the source of a bloodstream infection observed in more than 50 South American cancer patients.
Using whole-genome sequence typing (WGST), the team was able to link infection with the fungus Sarocladium kiliense to a tainted antinausea medication, ondansetron, that was given to cancer patients in Chile and Colombia.
This work is described in Emerging Infectious Diseases.
“Contamination of medical products, particularly with environmental fungi, poses growing concern and a public health threat, especially in vulnerable populations such as cancer patients,” said study author David Engelthaler, PhD, of The Translational Genomics Research Institute in Flagstaff, Arizona.
“Increased vigilance and the use of advanced technologies are needed to rapidly identify the likely sources of infection to efficiently guide epidemiologic investigations and initiate appropriate control measures.”
The S kiliense bloodstream-infection outbreak, which occurred from June 2013 through January 2014, included a cluster of cases at 8 hospitals in Santiago, Chile.
All of the patients received the same 4 intravenous medications. But only the antinausea medication ondansetron was given exclusively to cancer patients.
All of the patients infected with S kiliense received ondansetron from the same source, a pharmaceutical company called Vitrofarma SA (specifically, Plant No. 8 in Bogotá, Colombia). The drug was imported by LabVitales Chile SA and distributed by Pharma Isa Ltda.
Two of 3 lots of unopened ondansetron, tested by the Chilean Ministry of Health, yielded vials contaminated with S kiliense, forcing a recall of all ondansetron in Chile that was made by Vitrofarma SA.
Subsequently, Colombian officials discovered 14 other cases in which patients who were given ondansetron from Vitrofarma SA were infected with S kiliense.
S kiliense has been implicated in healthcare-related infections before, but the lack of available typing methods has precluded the ability to substantiate sources.
“The use of WGST to investigate fungal outbreaks has become integral to epidemiologic investigations,” Dr Engelthaler said. “Our WGST analysis demonstrated that the patient isolates from Chile and Colombia were nearly genetically indistinguishable from those recovered from the unopened medication vials, indicating the likely presence of a single-source infection.”