User login
Team identifies problems with preclinical research
Poor study design and the tendency to publish positive—but not negative—results threaten the validity of preclinical research, according to an article published in eLife.
“Only a fraction of drugs that show promise in animals end up proving safe and effective in humans,” said study author Jonathan Kimmelman, PhD, of McGill University in Montreal, Quebec, Canada.
“An important reason is because studies in animals are often not well designed and because positive results have a higher chance of being published. They end up skewing what we think we know about the potential of a drug.”
Dr Kimmelman and his colleagues came to this conclusion after evaluating all published animal studies of sunitinib, a drug used to treat advanced kidney cancer, a rare type of stomach cancer, and rare tumors of the neuroendocrine system.
The investigators found evidence to suggest that studies reporting little or no anticancer effect were simply not published, leading anticancer effects of the drug to be overestimated by as much as 45%.
The team noted, however, that these findings do not raise any concerns about the clinical use of sunitinib.
Dr Kimmelman and his colleagues also found that few studies used practices like blinding or randomization. And it was often unclear how many animals had been tested because the sample size was not reported.
The drug was tested against different cancers, and all types tested showed statistically significant anticancer activity, a result that “strains credibility”, according to Dr Kimmelman.
He added that researchers failed to observe the dose-dependent response to the drug that is known to occur in humans.
Finally, the researchers did not test the drug on a range of animal models, focusing instead on juvenile female mice with a compromised immune system. Malignancies tested in a wider range of animal types, such as mice that have spontaneously developed tumors, showed less extreme effect sizes.
“Preclinical research is plagued by poor design and reporting practices, exposing patients to harmful and inactive agents, wasting time in the lab, and driving up the price of drugs,” Dr Kimmelman said.
“Our findings provide compelling reasons for developing and implementing guidelines for the design and reporting of preclinical studies in cancer, similar to those already in use for stroke, epilepsy, and cardiology.” ![]()

Poor study design and the tendency to publish positive—but not negative—results threaten the validity of preclinical research, according to an article published in eLife.
“Only a fraction of drugs that show promise in animals end up proving safe and effective in humans,” said study author Jonathan Kimmelman, PhD, of McGill University in Montreal, Quebec, Canada.
“An important reason is because studies in animals are often not well designed and because positive results have a higher chance of being published. They end up skewing what we think we know about the potential of a drug.”
Dr Kimmelman and his colleagues came to this conclusion after evaluating all published animal studies of sunitinib, a drug used to treat advanced kidney cancer, a rare type of stomach cancer, and rare tumors of the neuroendocrine system.
The investigators found evidence to suggest that studies reporting little or no anticancer effect were simply not published, leading anticancer effects of the drug to be overestimated by as much as 45%.
The team noted, however, that these findings do not raise any concerns about the clinical use of sunitinib.
Dr Kimmelman and his colleagues also found that few studies used practices like blinding or randomization. And it was often unclear how many animals had been tested because the sample size was not reported.
The drug was tested against different cancers, and all types tested showed statistically significant anticancer activity, a result that “strains credibility”, according to Dr Kimmelman.
He added that researchers failed to observe the dose-dependent response to the drug that is known to occur in humans.
Finally, the researchers did not test the drug on a range of animal models, focusing instead on juvenile female mice with a compromised immune system. Malignancies tested in a wider range of animal types, such as mice that have spontaneously developed tumors, showed less extreme effect sizes.
“Preclinical research is plagued by poor design and reporting practices, exposing patients to harmful and inactive agents, wasting time in the lab, and driving up the price of drugs,” Dr Kimmelman said.
“Our findings provide compelling reasons for developing and implementing guidelines for the design and reporting of preclinical studies in cancer, similar to those already in use for stroke, epilepsy, and cardiology.” ![]()

Poor study design and the tendency to publish positive—but not negative—results threaten the validity of preclinical research, according to an article published in eLife.
“Only a fraction of drugs that show promise in animals end up proving safe and effective in humans,” said study author Jonathan Kimmelman, PhD, of McGill University in Montreal, Quebec, Canada.
“An important reason is because studies in animals are often not well designed and because positive results have a higher chance of being published. They end up skewing what we think we know about the potential of a drug.”
Dr Kimmelman and his colleagues came to this conclusion after evaluating all published animal studies of sunitinib, a drug used to treat advanced kidney cancer, a rare type of stomach cancer, and rare tumors of the neuroendocrine system.
The investigators found evidence to suggest that studies reporting little or no anticancer effect were simply not published, leading anticancer effects of the drug to be overestimated by as much as 45%.
The team noted, however, that these findings do not raise any concerns about the clinical use of sunitinib.
Dr Kimmelman and his colleagues also found that few studies used practices like blinding or randomization. And it was often unclear how many animals had been tested because the sample size was not reported.
The drug was tested against different cancers, and all types tested showed statistically significant anticancer activity, a result that “strains credibility”, according to Dr Kimmelman.
He added that researchers failed to observe the dose-dependent response to the drug that is known to occur in humans.
Finally, the researchers did not test the drug on a range of animal models, focusing instead on juvenile female mice with a compromised immune system. Malignancies tested in a wider range of animal types, such as mice that have spontaneously developed tumors, showed less extreme effect sizes.
“Preclinical research is plagued by poor design and reporting practices, exposing patients to harmful and inactive agents, wasting time in the lab, and driving up the price of drugs,” Dr Kimmelman said.
“Our findings provide compelling reasons for developing and implementing guidelines for the design and reporting of preclinical studies in cancer, similar to those already in use for stroke, epilepsy, and cardiology.” ![]()
NCCN creates tool to aid treatment decisions

patient and her father
Photo by Rhoda Baer
The National Comprehensive Cancer Network (NCCN) has developed a new tool to accompany its clinical practice guidelines.
The tool—known as NCCN Evidence Blocks™—is designed to provide additional information about guideline recommendations and help inform treatment decisions.
NCCN has already added Evidence Blocks to its guidelines for chronic myelogenous leukemia and multiple myeloma.
The organization hopes to have Evidence Blocks for all of its guidelines by early 2017.
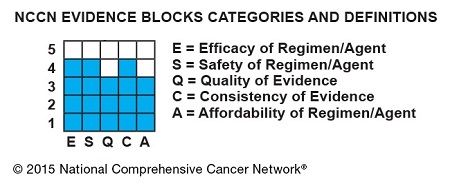
The Evidence Blocks provide a visual representation of 5 key value measures pertaining to guideline recommendations:
- Efficacy of treatment regimens
- Safety of regimens
- Quality and quantity of evidence supporting regimens
- Consistency of evidence supporting regimens
- Affordability of regimens. (This represents an estimate of overall total cost of a therapy, including but not limited to acquisition, administration, inpatient vs outpatient care, supportive care, infusions, toxicity monitoring, antiemetics and growth factors, and hospitalization.)
The Evidence Blocks are graphics of actual blocks that consist of 25 small squares. So each block has 5 rows and 5 columns.
Each of the 5 value measures—efficacy, safety, etc.—has a dedicated column within an Evidence Block, and each row of the Evidence Block represents a rating on a scale of 1 to 5. A score of 1 is unfavorable and a score of 5 is most favorable.
The rating of each value measure is shown by filling in the squares of the dedicated column—such as efficacy—up to the row that represents its assigned score—such as 4.
For example:
NCCN hopes this visual rating system will help patients and their physicians identify the optimal treatment based on clinical and economic considerations that are of the most value to the patient.
“Some patients will want an emerging therapy even with limited data,” said Robert W. Carlson, MD, chief executive officer of NCCN.
“Others will be most concerned about the expected side effects of the treatment indicated in the safety column. Still others may be very sensitive to cost. By considering the attributes of the range of possible therapies, the healthcare provider and the patient can discuss the benefits and drawbacks of each option and come to a decision most acceptable to the individual.”
By the end of 2015, NCCN expects to publish NCCN Evidence Blocks for systemic therapies (not surgery or radiation therapy) in the NCCN guidelines for breast, colon, non-small cell lung, and rectal cancers.
NCCN Evidence Blocks for systemic therapies are expected to be contained within the complete library of NCCN guidelines by the end of 2016.
In the near term, NCCN will continue to publish 2 sets of guidelines: those including NCCN Evidence Blocks and those without. The Evidence Blocks are not currently published in the NCCN Guidelines for Patients® and are intended for use in the US only.
For more information about NCCN Evidence Blocks, visit NCCN.org/EvidenceBlocks. ![]()

patient and her father
Photo by Rhoda Baer
The National Comprehensive Cancer Network (NCCN) has developed a new tool to accompany its clinical practice guidelines.
The tool—known as NCCN Evidence Blocks™—is designed to provide additional information about guideline recommendations and help inform treatment decisions.
NCCN has already added Evidence Blocks to its guidelines for chronic myelogenous leukemia and multiple myeloma.
The organization hopes to have Evidence Blocks for all of its guidelines by early 2017.
The Evidence Blocks provide a visual representation of 5 key value measures pertaining to guideline recommendations:
- Efficacy of treatment regimens
- Safety of regimens
- Quality and quantity of evidence supporting regimens
- Consistency of evidence supporting regimens
- Affordability of regimens. (This represents an estimate of overall total cost of a therapy, including but not limited to acquisition, administration, inpatient vs outpatient care, supportive care, infusions, toxicity monitoring, antiemetics and growth factors, and hospitalization.)
The Evidence Blocks are graphics of actual blocks that consist of 25 small squares. So each block has 5 rows and 5 columns.
Each of the 5 value measures—efficacy, safety, etc.—has a dedicated column within an Evidence Block, and each row of the Evidence Block represents a rating on a scale of 1 to 5. A score of 1 is unfavorable and a score of 5 is most favorable.
The rating of each value measure is shown by filling in the squares of the dedicated column—such as efficacy—up to the row that represents its assigned score—such as 4.
For example:

NCCN hopes this visual rating system will help patients and their physicians identify the optimal treatment based on clinical and economic considerations that are of the most value to the patient.
“Some patients will want an emerging therapy even with limited data,” said Robert W. Carlson, MD, chief executive officer of NCCN.
“Others will be most concerned about the expected side effects of the treatment indicated in the safety column. Still others may be very sensitive to cost. By considering the attributes of the range of possible therapies, the healthcare provider and the patient can discuss the benefits and drawbacks of each option and come to a decision most acceptable to the individual.”
By the end of 2015, NCCN expects to publish NCCN Evidence Blocks for systemic therapies (not surgery or radiation therapy) in the NCCN guidelines for breast, colon, non-small cell lung, and rectal cancers.
NCCN Evidence Blocks for systemic therapies are expected to be contained within the complete library of NCCN guidelines by the end of 2016.
In the near term, NCCN will continue to publish 2 sets of guidelines: those including NCCN Evidence Blocks and those without. The Evidence Blocks are not currently published in the NCCN Guidelines for Patients® and are intended for use in the US only.
For more information about NCCN Evidence Blocks, visit NCCN.org/EvidenceBlocks. ![]()

patient and her father
Photo by Rhoda Baer
The National Comprehensive Cancer Network (NCCN) has developed a new tool to accompany its clinical practice guidelines.
The tool—known as NCCN Evidence Blocks™—is designed to provide additional information about guideline recommendations and help inform treatment decisions.
NCCN has already added Evidence Blocks to its guidelines for chronic myelogenous leukemia and multiple myeloma.
The organization hopes to have Evidence Blocks for all of its guidelines by early 2017.
The Evidence Blocks provide a visual representation of 5 key value measures pertaining to guideline recommendations:
- Efficacy of treatment regimens
- Safety of regimens
- Quality and quantity of evidence supporting regimens
- Consistency of evidence supporting regimens
- Affordability of regimens. (This represents an estimate of overall total cost of a therapy, including but not limited to acquisition, administration, inpatient vs outpatient care, supportive care, infusions, toxicity monitoring, antiemetics and growth factors, and hospitalization.)
The Evidence Blocks are graphics of actual blocks that consist of 25 small squares. So each block has 5 rows and 5 columns.
Each of the 5 value measures—efficacy, safety, etc.—has a dedicated column within an Evidence Block, and each row of the Evidence Block represents a rating on a scale of 1 to 5. A score of 1 is unfavorable and a score of 5 is most favorable.
The rating of each value measure is shown by filling in the squares of the dedicated column—such as efficacy—up to the row that represents its assigned score—such as 4.
For example:

NCCN hopes this visual rating system will help patients and their physicians identify the optimal treatment based on clinical and economic considerations that are of the most value to the patient.
“Some patients will want an emerging therapy even with limited data,” said Robert W. Carlson, MD, chief executive officer of NCCN.
“Others will be most concerned about the expected side effects of the treatment indicated in the safety column. Still others may be very sensitive to cost. By considering the attributes of the range of possible therapies, the healthcare provider and the patient can discuss the benefits and drawbacks of each option and come to a decision most acceptable to the individual.”
By the end of 2015, NCCN expects to publish NCCN Evidence Blocks for systemic therapies (not surgery or radiation therapy) in the NCCN guidelines for breast, colon, non-small cell lung, and rectal cancers.
NCCN Evidence Blocks for systemic therapies are expected to be contained within the complete library of NCCN guidelines by the end of 2016.
In the near term, NCCN will continue to publish 2 sets of guidelines: those including NCCN Evidence Blocks and those without. The Evidence Blocks are not currently published in the NCCN Guidelines for Patients® and are intended for use in the US only.
For more information about NCCN Evidence Blocks, visit NCCN.org/EvidenceBlocks. ![]()
Malarial protein is basis for potential cancer therapy

Photo by Vivian Sum
A protein expressed by the malaria parasite Plasmodium falciparum may prove useful for treating a range of cancers, according to research published in Cancer Cell.
This protein, VAR2CSA, binds a chondroitin sulfate (CS) that is found in placenta but is also present in many different cancer cells.
So investigators combined recombinant VAR2CSA (rVAR2) with 2 different toxins to create cancer-targeting treatments.
The treatments effectively targeted cancers in vitro and in vivo, impeding tumor growth and even eradicating cancer in some mice.
An idea is born
This research was born while the investigators were exploring why pregnant women are particularly susceptible to malaria. The team found that P falciparum produces VAR2CSA, which binds to a particular CS in the placenta, and that placenta-like CS (pl-CS) is found in most cancers.
This suggested the pl-CS could be a target for anticancer drugs, and VAR2CSA could provide the tool for carrying such drugs to tumors.
“Scientists have spent decades trying to find biochemical similarities between placenta tissue and cancer, but we just didn’t have the technology to find it,” said project leader Mads Daugaard, PhD, of the University of British Columbia in Vancouver, Canada.
“When my colleagues discovered how malaria uses VAR2CSA to embed itself in the placenta, we immediately saw its potential to deliver cancer drugs in a precise, controlled way to tumors.”
Testing rVAR2
After demonstrating that rVAR2 binds only to pl-CS, the investigators tested rVAR2 in patient-derived cancer cell lines of hematopoietic, epithelial, and mesenchymal origin. rVAR2 reacted with 95% (106/111) of these cell lines.
To determine whether rVAR2 could be used as a pl-CS-specific tumor-targeting system, the investigators fused the cytotoxic domain of diphtheria toxin (DT388) to rVAR2, creating a recombinant rDT388-VAR2 (rVAR2-DT) fusion protein.
The rVAR2-DT protein killed tumor cell lines of epithelial and mesenchymal origin, but it had no effect on normal primary human endothelial cells.
The investigators also tested rVAR2-DT in mouse models of prostate cancer and found that as few as 3 doses were enough to significantly inhibit tumor growth.
However, the team noted that clinical trials with DT fusions have shown that high drug concentrations are not well-tolerated.
So they chemically conjugated a hemiasterlin analog (KT886) to rVAR2 via a protease cleavable linker. The rVAR2-KT886 drug conjugate (VDC886) carried an average of 3 toxins per rVAR2 molecule.
The investigators tested VDC886 in 33 cancer cell lines and observed cytotoxicity in all cell lines.
So the team went on to test VDC886 in mouse models of non-Hodgkin lymphoma, prostate cancer, and metastatic breast cancer. VDC886 significantly inhibited tumor growth in all 3 models.
In mice with non-Hodgkin lymphoma, treated tumors were about a quarter the size of tumors in control mice. For the mice with prostate cancer, tumors completely disappeared in 2 of the 6 treated mice a month after they received the first dose of VDC886.
In mice with metastatic breast cancer, 5 of the 6 treated mice were cured and alive after almost 8 weeks. None of the control mice with metastatic breast cancer survived that long.
The investigators said they did not observe any adverse effects in the mice, and their organs were unharmed by the therapy.
“It appears that the malaria protein attaches itself to the tumor without any significant attachment to other tissue,” said Thomas Mandel Clausen, a PhD student at the University of Copenhagen in Denmark.
“And the mice that were given doses of protein and toxin showed far higher survival rates than the untreated mice. We have seen that 3 doses can arrest growth in a tumor and even make it shrink.”
Based on these results, 2 companies—Vancouver-based Kairos Therapeutics and Copenhagen-based VAR2 Pharmaceuticals—are developing the compound for clinical trials. The investigators believe this will take a few years. ![]()

Photo by Vivian Sum
A protein expressed by the malaria parasite Plasmodium falciparum may prove useful for treating a range of cancers, according to research published in Cancer Cell.
This protein, VAR2CSA, binds a chondroitin sulfate (CS) that is found in placenta but is also present in many different cancer cells.
So investigators combined recombinant VAR2CSA (rVAR2) with 2 different toxins to create cancer-targeting treatments.
The treatments effectively targeted cancers in vitro and in vivo, impeding tumor growth and even eradicating cancer in some mice.
An idea is born
This research was born while the investigators were exploring why pregnant women are particularly susceptible to malaria. The team found that P falciparum produces VAR2CSA, which binds to a particular CS in the placenta, and that placenta-like CS (pl-CS) is found in most cancers.
This suggested the pl-CS could be a target for anticancer drugs, and VAR2CSA could provide the tool for carrying such drugs to tumors.
“Scientists have spent decades trying to find biochemical similarities between placenta tissue and cancer, but we just didn’t have the technology to find it,” said project leader Mads Daugaard, PhD, of the University of British Columbia in Vancouver, Canada.
“When my colleagues discovered how malaria uses VAR2CSA to embed itself in the placenta, we immediately saw its potential to deliver cancer drugs in a precise, controlled way to tumors.”
Testing rVAR2
After demonstrating that rVAR2 binds only to pl-CS, the investigators tested rVAR2 in patient-derived cancer cell lines of hematopoietic, epithelial, and mesenchymal origin. rVAR2 reacted with 95% (106/111) of these cell lines.
To determine whether rVAR2 could be used as a pl-CS-specific tumor-targeting system, the investigators fused the cytotoxic domain of diphtheria toxin (DT388) to rVAR2, creating a recombinant rDT388-VAR2 (rVAR2-DT) fusion protein.
The rVAR2-DT protein killed tumor cell lines of epithelial and mesenchymal origin, but it had no effect on normal primary human endothelial cells.
The investigators also tested rVAR2-DT in mouse models of prostate cancer and found that as few as 3 doses were enough to significantly inhibit tumor growth.
However, the team noted that clinical trials with DT fusions have shown that high drug concentrations are not well-tolerated.
So they chemically conjugated a hemiasterlin analog (KT886) to rVAR2 via a protease cleavable linker. The rVAR2-KT886 drug conjugate (VDC886) carried an average of 3 toxins per rVAR2 molecule.
The investigators tested VDC886 in 33 cancer cell lines and observed cytotoxicity in all cell lines.
So the team went on to test VDC886 in mouse models of non-Hodgkin lymphoma, prostate cancer, and metastatic breast cancer. VDC886 significantly inhibited tumor growth in all 3 models.
In mice with non-Hodgkin lymphoma, treated tumors were about a quarter the size of tumors in control mice. For the mice with prostate cancer, tumors completely disappeared in 2 of the 6 treated mice a month after they received the first dose of VDC886.
In mice with metastatic breast cancer, 5 of the 6 treated mice were cured and alive after almost 8 weeks. None of the control mice with metastatic breast cancer survived that long.
The investigators said they did not observe any adverse effects in the mice, and their organs were unharmed by the therapy.
“It appears that the malaria protein attaches itself to the tumor without any significant attachment to other tissue,” said Thomas Mandel Clausen, a PhD student at the University of Copenhagen in Denmark.
“And the mice that were given doses of protein and toxin showed far higher survival rates than the untreated mice. We have seen that 3 doses can arrest growth in a tumor and even make it shrink.”
Based on these results, 2 companies—Vancouver-based Kairos Therapeutics and Copenhagen-based VAR2 Pharmaceuticals—are developing the compound for clinical trials. The investigators believe this will take a few years. ![]()

Photo by Vivian Sum
A protein expressed by the malaria parasite Plasmodium falciparum may prove useful for treating a range of cancers, according to research published in Cancer Cell.
This protein, VAR2CSA, binds a chondroitin sulfate (CS) that is found in placenta but is also present in many different cancer cells.
So investigators combined recombinant VAR2CSA (rVAR2) with 2 different toxins to create cancer-targeting treatments.
The treatments effectively targeted cancers in vitro and in vivo, impeding tumor growth and even eradicating cancer in some mice.
An idea is born
This research was born while the investigators were exploring why pregnant women are particularly susceptible to malaria. The team found that P falciparum produces VAR2CSA, which binds to a particular CS in the placenta, and that placenta-like CS (pl-CS) is found in most cancers.
This suggested the pl-CS could be a target for anticancer drugs, and VAR2CSA could provide the tool for carrying such drugs to tumors.
“Scientists have spent decades trying to find biochemical similarities between placenta tissue and cancer, but we just didn’t have the technology to find it,” said project leader Mads Daugaard, PhD, of the University of British Columbia in Vancouver, Canada.
“When my colleagues discovered how malaria uses VAR2CSA to embed itself in the placenta, we immediately saw its potential to deliver cancer drugs in a precise, controlled way to tumors.”
Testing rVAR2
After demonstrating that rVAR2 binds only to pl-CS, the investigators tested rVAR2 in patient-derived cancer cell lines of hematopoietic, epithelial, and mesenchymal origin. rVAR2 reacted with 95% (106/111) of these cell lines.
To determine whether rVAR2 could be used as a pl-CS-specific tumor-targeting system, the investigators fused the cytotoxic domain of diphtheria toxin (DT388) to rVAR2, creating a recombinant rDT388-VAR2 (rVAR2-DT) fusion protein.
The rVAR2-DT protein killed tumor cell lines of epithelial and mesenchymal origin, but it had no effect on normal primary human endothelial cells.
The investigators also tested rVAR2-DT in mouse models of prostate cancer and found that as few as 3 doses were enough to significantly inhibit tumor growth.
However, the team noted that clinical trials with DT fusions have shown that high drug concentrations are not well-tolerated.
So they chemically conjugated a hemiasterlin analog (KT886) to rVAR2 via a protease cleavable linker. The rVAR2-KT886 drug conjugate (VDC886) carried an average of 3 toxins per rVAR2 molecule.
The investigators tested VDC886 in 33 cancer cell lines and observed cytotoxicity in all cell lines.
So the team went on to test VDC886 in mouse models of non-Hodgkin lymphoma, prostate cancer, and metastatic breast cancer. VDC886 significantly inhibited tumor growth in all 3 models.
In mice with non-Hodgkin lymphoma, treated tumors were about a quarter the size of tumors in control mice. For the mice with prostate cancer, tumors completely disappeared in 2 of the 6 treated mice a month after they received the first dose of VDC886.
In mice with metastatic breast cancer, 5 of the 6 treated mice were cured and alive after almost 8 weeks. None of the control mice with metastatic breast cancer survived that long.
The investigators said they did not observe any adverse effects in the mice, and their organs were unharmed by the therapy.
“It appears that the malaria protein attaches itself to the tumor without any significant attachment to other tissue,” said Thomas Mandel Clausen, a PhD student at the University of Copenhagen in Denmark.
“And the mice that were given doses of protein and toxin showed far higher survival rates than the untreated mice. We have seen that 3 doses can arrest growth in a tumor and even make it shrink.”
Based on these results, 2 companies—Vancouver-based Kairos Therapeutics and Copenhagen-based VAR2 Pharmaceuticals—are developing the compound for clinical trials. The investigators believe this will take a few years. ![]()
Smartphone use may put patient data at risk

Photo by Daniel Sone
A new survey suggests that doctors and nurses in London are routinely using their own smartphones for patient care.
Investigators say the current lack of data encryption on these devices could result in the inadvertent disclosure of “highly sensitive and confidential data” in the absence of an active organizational strategy on digital security.
The team raised this issue and reported results of the survey in BMJ Innovations.
Mohammad H. Mobasheri, MBBS, of Imperial College London in the UK, and his colleagues wanted to determine how healthcare professionals are using digital technology on the frontline.
So the investigators invited more than 6000 clinical staff at 5 London hospitals of varying sizes to complete a questionnaire on ownership and use of portable devices and mobile health apps in the workplace.
The results are based on the responses of 287 doctors and 564 nurses from different specialties.
About 99% of doctors said they owned a smartphone, and 73.5% owned a tablet. The equivalent figures for nurses were 95.1% and 64.7%, respectively.
When asked about the usefulness of smartphones for carrying out clinical duties, 92.6% of doctors and 53.2% of nurses said these devices were “very useful” or “useful.”
Most doctors (93.8%) used their smartphone while at work to communicate with their colleagues, compared with 28.5% of nurses. About half of the doctors (50.2%) used their smartphone instead of a traditional bleep (page).
About 78% of doctors and 34.8% of nurses said they had downloaded a medical app to their device, with 89.6% of doctors and 67.1% of nurses saying they used these apps as part of their clinical work.
Of those who owned a medical app and used it at work, 41.3% of doctors reported using such an app weekly, and 33% said they used one daily. The equivalent figures for nurses were 42% and 22.3%, respectively.
The apps included drug formularies, medical calculators, and those for disease diagnosis and treatment, reference and education, documentation and drug preparation.
When asked if they had ever sent patient data over their smartphones using SMS, app-based messaging (such as WhatsApp), and picture messaging using their smartphone camera, many respondents said they had done so.
About 65% of doctors had used SMS, 33.1% had used app-based messaging, and 46% had used their phone’s camera and picture messaging to send a photo of a wound or X-ray to a colleague. The corresponding figures for nurses were much lower—13.8%, 5.7%, and 7.4%, respectively.
About 28% of doctors and 3.6% of nurses said they still retained clinical information on their smartphones.
A substantial proportion of respondents said they wanted to be able to use their own devices at work. About 72% of doctors and 37.2% of nurses wanted a secure means of sending patient data to colleagues using their own smartphone.
Fully secure messaging services for smartphones are not yet available in the UK, and the data are unlikely to be encrypted, according to investigators. They therefore urged National Health Service organizations to make sure their staff understands the potential risks of sharing patient information via their unsecured smartphones.
The team said the results of this survey provide strong evidence that healthcare organizations need to develop policies to support the safe and secure use of digital technologies in the workplace. ![]()

Photo by Daniel Sone
A new survey suggests that doctors and nurses in London are routinely using their own smartphones for patient care.
Investigators say the current lack of data encryption on these devices could result in the inadvertent disclosure of “highly sensitive and confidential data” in the absence of an active organizational strategy on digital security.
The team raised this issue and reported results of the survey in BMJ Innovations.
Mohammad H. Mobasheri, MBBS, of Imperial College London in the UK, and his colleagues wanted to determine how healthcare professionals are using digital technology on the frontline.
So the investigators invited more than 6000 clinical staff at 5 London hospitals of varying sizes to complete a questionnaire on ownership and use of portable devices and mobile health apps in the workplace.
The results are based on the responses of 287 doctors and 564 nurses from different specialties.
About 99% of doctors said they owned a smartphone, and 73.5% owned a tablet. The equivalent figures for nurses were 95.1% and 64.7%, respectively.
When asked about the usefulness of smartphones for carrying out clinical duties, 92.6% of doctors and 53.2% of nurses said these devices were “very useful” or “useful.”
Most doctors (93.8%) used their smartphone while at work to communicate with their colleagues, compared with 28.5% of nurses. About half of the doctors (50.2%) used their smartphone instead of a traditional bleep (page).
About 78% of doctors and 34.8% of nurses said they had downloaded a medical app to their device, with 89.6% of doctors and 67.1% of nurses saying they used these apps as part of their clinical work.
Of those who owned a medical app and used it at work, 41.3% of doctors reported using such an app weekly, and 33% said they used one daily. The equivalent figures for nurses were 42% and 22.3%, respectively.
The apps included drug formularies, medical calculators, and those for disease diagnosis and treatment, reference and education, documentation and drug preparation.
When asked if they had ever sent patient data over their smartphones using SMS, app-based messaging (such as WhatsApp), and picture messaging using their smartphone camera, many respondents said they had done so.
About 65% of doctors had used SMS, 33.1% had used app-based messaging, and 46% had used their phone’s camera and picture messaging to send a photo of a wound or X-ray to a colleague. The corresponding figures for nurses were much lower—13.8%, 5.7%, and 7.4%, respectively.
About 28% of doctors and 3.6% of nurses said they still retained clinical information on their smartphones.
A substantial proportion of respondents said they wanted to be able to use their own devices at work. About 72% of doctors and 37.2% of nurses wanted a secure means of sending patient data to colleagues using their own smartphone.
Fully secure messaging services for smartphones are not yet available in the UK, and the data are unlikely to be encrypted, according to investigators. They therefore urged National Health Service organizations to make sure their staff understands the potential risks of sharing patient information via their unsecured smartphones.
The team said the results of this survey provide strong evidence that healthcare organizations need to develop policies to support the safe and secure use of digital technologies in the workplace. ![]()

Photo by Daniel Sone
A new survey suggests that doctors and nurses in London are routinely using their own smartphones for patient care.
Investigators say the current lack of data encryption on these devices could result in the inadvertent disclosure of “highly sensitive and confidential data” in the absence of an active organizational strategy on digital security.
The team raised this issue and reported results of the survey in BMJ Innovations.
Mohammad H. Mobasheri, MBBS, of Imperial College London in the UK, and his colleagues wanted to determine how healthcare professionals are using digital technology on the frontline.
So the investigators invited more than 6000 clinical staff at 5 London hospitals of varying sizes to complete a questionnaire on ownership and use of portable devices and mobile health apps in the workplace.
The results are based on the responses of 287 doctors and 564 nurses from different specialties.
About 99% of doctors said they owned a smartphone, and 73.5% owned a tablet. The equivalent figures for nurses were 95.1% and 64.7%, respectively.
When asked about the usefulness of smartphones for carrying out clinical duties, 92.6% of doctors and 53.2% of nurses said these devices were “very useful” or “useful.”
Most doctors (93.8%) used their smartphone while at work to communicate with their colleagues, compared with 28.5% of nurses. About half of the doctors (50.2%) used their smartphone instead of a traditional bleep (page).
About 78% of doctors and 34.8% of nurses said they had downloaded a medical app to their device, with 89.6% of doctors and 67.1% of nurses saying they used these apps as part of their clinical work.
Of those who owned a medical app and used it at work, 41.3% of doctors reported using such an app weekly, and 33% said they used one daily. The equivalent figures for nurses were 42% and 22.3%, respectively.
The apps included drug formularies, medical calculators, and those for disease diagnosis and treatment, reference and education, documentation and drug preparation.
When asked if they had ever sent patient data over their smartphones using SMS, app-based messaging (such as WhatsApp), and picture messaging using their smartphone camera, many respondents said they had done so.
About 65% of doctors had used SMS, 33.1% had used app-based messaging, and 46% had used their phone’s camera and picture messaging to send a photo of a wound or X-ray to a colleague. The corresponding figures for nurses were much lower—13.8%, 5.7%, and 7.4%, respectively.
About 28% of doctors and 3.6% of nurses said they still retained clinical information on their smartphones.
A substantial proportion of respondents said they wanted to be able to use their own devices at work. About 72% of doctors and 37.2% of nurses wanted a secure means of sending patient data to colleagues using their own smartphone.
Fully secure messaging services for smartphones are not yet available in the UK, and the data are unlikely to be encrypted, according to investigators. They therefore urged National Health Service organizations to make sure their staff understands the potential risks of sharing patient information via their unsecured smartphones.
The team said the results of this survey provide strong evidence that healthcare organizations need to develop policies to support the safe and secure use of digital technologies in the workplace. ![]()
Trio wins Nobel Prize for DNA repair discoveries
Image by Tom Ellenberger
Three researchers have won this year’s Nobel Prize in Chemistry for mechanistic studies of DNA repair.
Tomas Lindahl, MD, PhD, Paul Modrich, PhD, and Aziz Sancar, MD, PhD, each mapped how DNA repair systems function at a detailed molecular level.
Their work has provided insight into how cells function, knowledge that can be used in the development of new cancer treatments, among other applications.
In the early 1970s, scientists believed that DNA was an extremely stable molecule, but Dr Lindahl demonstrated that DNA decays at a rate that ought to have made life on Earth impossible.
This insight led to the discovery of molecular machinery known as base excision repair, which constantly counteracts the collapse of our DNA.
For his part, Dr Sancar mapped nucleotide excision repair, the mechanism that cells use to repair UV damage to DNA.
People born with defects in this repair system will develop skin cancer if they are exposed to sunlight. The cell also utilizes nucleotide excision repair to correct defects caused by mutagenic substances, among other things.
Dr Modrich demonstrated how the cell corrects errors that occur when DNA is replicated during cell division.
This mechanism, mismatch repair, reduces the error frequency during DNA replication by about a thousand-fold. Congenital defects in mismatch repair are known, for example, to cause a hereditary variant of colon cancer.
About the winners
Tomas Lindahl was born in 1938 in Stockholm, Sweden. He earned his PhD in 1967 and his MD in 1970, both from Karolinska Institutet in Sweden. He is currently emeritus group leader at the Francis Crick Institute in London, UK.
Paul Modrich was born in 1946. In 1973, he earned his PhD from Stanford University in California. He is currently an investigator at Howard Hughes Medical Institute in Chevy Chase, Maryland, and a professor at Duke University School of Medicine in Durham, North Carolina.
Aziz Sancar was born in 1946 in Savur, Turkey. He earned his MD in 1969 from Istanbul University in Turkey and his PhD in 1977 from the University of Texas in Dallas. He is currently a professor at the University of North Carolina School of Medicine in Chapel Hill. ![]()

Image by Tom Ellenberger
Three researchers have won this year’s Nobel Prize in Chemistry for mechanistic studies of DNA repair.
Tomas Lindahl, MD, PhD, Paul Modrich, PhD, and Aziz Sancar, MD, PhD, each mapped how DNA repair systems function at a detailed molecular level.
Their work has provided insight into how cells function, knowledge that can be used in the development of new cancer treatments, among other applications.
In the early 1970s, scientists believed that DNA was an extremely stable molecule, but Dr Lindahl demonstrated that DNA decays at a rate that ought to have made life on Earth impossible.
This insight led to the discovery of molecular machinery known as base excision repair, which constantly counteracts the collapse of our DNA.
For his part, Dr Sancar mapped nucleotide excision repair, the mechanism that cells use to repair UV damage to DNA.
People born with defects in this repair system will develop skin cancer if they are exposed to sunlight. The cell also utilizes nucleotide excision repair to correct defects caused by mutagenic substances, among other things.
Dr Modrich demonstrated how the cell corrects errors that occur when DNA is replicated during cell division.
This mechanism, mismatch repair, reduces the error frequency during DNA replication by about a thousand-fold. Congenital defects in mismatch repair are known, for example, to cause a hereditary variant of colon cancer.
About the winners
Tomas Lindahl was born in 1938 in Stockholm, Sweden. He earned his PhD in 1967 and his MD in 1970, both from Karolinska Institutet in Sweden. He is currently emeritus group leader at the Francis Crick Institute in London, UK.
Paul Modrich was born in 1946. In 1973, he earned his PhD from Stanford University in California. He is currently an investigator at Howard Hughes Medical Institute in Chevy Chase, Maryland, and a professor at Duke University School of Medicine in Durham, North Carolina.
Aziz Sancar was born in 1946 in Savur, Turkey. He earned his MD in 1969 from Istanbul University in Turkey and his PhD in 1977 from the University of Texas in Dallas. He is currently a professor at the University of North Carolina School of Medicine in Chapel Hill. ![]()

Image by Tom Ellenberger
Three researchers have won this year’s Nobel Prize in Chemistry for mechanistic studies of DNA repair.
Tomas Lindahl, MD, PhD, Paul Modrich, PhD, and Aziz Sancar, MD, PhD, each mapped how DNA repair systems function at a detailed molecular level.
Their work has provided insight into how cells function, knowledge that can be used in the development of new cancer treatments, among other applications.
In the early 1970s, scientists believed that DNA was an extremely stable molecule, but Dr Lindahl demonstrated that DNA decays at a rate that ought to have made life on Earth impossible.
This insight led to the discovery of molecular machinery known as base excision repair, which constantly counteracts the collapse of our DNA.
For his part, Dr Sancar mapped nucleotide excision repair, the mechanism that cells use to repair UV damage to DNA.
People born with defects in this repair system will develop skin cancer if they are exposed to sunlight. The cell also utilizes nucleotide excision repair to correct defects caused by mutagenic substances, among other things.
Dr Modrich demonstrated how the cell corrects errors that occur when DNA is replicated during cell division.
This mechanism, mismatch repair, reduces the error frequency during DNA replication by about a thousand-fold. Congenital defects in mismatch repair are known, for example, to cause a hereditary variant of colon cancer.
About the winners
Tomas Lindahl was born in 1938 in Stockholm, Sweden. He earned his PhD in 1967 and his MD in 1970, both from Karolinska Institutet in Sweden. He is currently emeritus group leader at the Francis Crick Institute in London, UK.
Paul Modrich was born in 1946. In 1973, he earned his PhD from Stanford University in California. He is currently an investigator at Howard Hughes Medical Institute in Chevy Chase, Maryland, and a professor at Duke University School of Medicine in Durham, North Carolina.
Aziz Sancar was born in 1946 in Savur, Turkey. He earned his MD in 1969 from Istanbul University in Turkey and his PhD in 1977 from the University of Texas in Dallas. He is currently a professor at the University of North Carolina School of Medicine in Chapel Hill. ![]()
Word choice affects public perception of drugs

Photo courtesy of the FDA
Using the words “breakthrough” and “promising” to describe new drugs affects the public’s perception of the drugs’ effectiveness, according to a study published in JAMA Internal Medicine.
Investigators noted that, in everyday usage, the term “breakthrough” represents a highly significant or definitive advance.
However, the US Food and Drug Administration’s (FDA’s) “breakthrough therapy designation” has a different meaning.
Since the Safety and Innovation Act became law in 2012, the FDA can assign breakthrough designation to a drug that “treats a serious or life-threatening condition” and “may demonstrate a substantial improvement . . . over available therapies” based on preliminary evidence.
And since the creation of the Safety and Innovation Act, all FDA press releases announcing the approval of breakthrough-designated drugs have used the term “breakthrough,” while about half have used the term “promising.”
“Today, patients and their families can easily find FDA press releases on the Internet, or they often hear about them in the news,” said study author Steven Woloshin, MD, of The Dartmouth Institute for Health Policy and Clinical Practice in Lebanon, New Hampshire.
“But the reality is that unless patients fully understand how the FDA is using the term ‘breakthrough,’ they may have unwarranted confidence in the evidence supporting drug claims. So we thought it was important to test how these terms affect the judgement of people without medical training.”
Survey details
The investigators conducted an online survey of 597 Americans. Participants were randomly given 1 of 5 short descriptions of a recently approved drug.
The descriptions were based on an FDA press release for a metastatic lung cancer breakthrough-designated drug that was conditionally approved based on the surrogate outcome of tumor shrinkage.
The first, “facts-only” description described the drug as meeting the criteria for breakthrough designation but did not actually use the term “breakthrough.”
A second and a third description included the facts and added the terms “breakthrough” and “promising,” respectively.
The fourth, “tentative” description included the facts, used the word “breakthrough,” and used the following FDA-required language for professional labeling:
The FDA pointed out that the drug was approved based on tumor shrinkage but that an improvement in survival or disease-related symptoms has not been established. Continued approval for this indication may be contingent upon verification and description of clinical benefit in confirmatory trials.
The fifth, “definitive” description included the same information as the tentative description, but “may be contingent” was changed to “is contingent.”
Participants were then asked to judge the drug’s benefit, harm, and strength of evidence.
Results
The investigators found that adding either “breakthrough” or “promising” in the description significantly increased the percentage of participants who rated the drug as “very” or “completely” effective compared with the facts-only description—23% and 25% vs 11%.
Adding “breakthrough” or “promising” to the description also significantly increased the number of people who reported believing that evidence supporting the drug is “strong” or “extremely strong”—59% and 63% vs 43%.
At the same time, adding either the tentative or definitive explanations significantly reduced the percentage of participants who believed (incorrectly) that the drug had been “proven to save lives”—16% tentative and 10% definitive vs 31% breakthrough.
Finally, when participants were asked which of 2 drugs—one described as “breakthrough,” the other as meeting the breakthrough criteria—they would take for a potentially deadly condition, 92% chose the “breakthrough” drug.
“Our findings clearly indicate that words like ‘breakthrough’ and ‘promising’ increase people’s beliefs in a drug’s effectiveness,” said Lisa Schwartz, MD, of The Dartmouth Institute for Health Policy and Clinical Practice.
“In light of [the findings], press releases with neutral terms and that clearly explain the limited evidence supporting what breakthrough designation and accelerated approval mean might help consumers make more accurate judgements about these drugs.” ![]()

Photo courtesy of the FDA
Using the words “breakthrough” and “promising” to describe new drugs affects the public’s perception of the drugs’ effectiveness, according to a study published in JAMA Internal Medicine.
Investigators noted that, in everyday usage, the term “breakthrough” represents a highly significant or definitive advance.
However, the US Food and Drug Administration’s (FDA’s) “breakthrough therapy designation” has a different meaning.
Since the Safety and Innovation Act became law in 2012, the FDA can assign breakthrough designation to a drug that “treats a serious or life-threatening condition” and “may demonstrate a substantial improvement . . . over available therapies” based on preliminary evidence.
And since the creation of the Safety and Innovation Act, all FDA press releases announcing the approval of breakthrough-designated drugs have used the term “breakthrough,” while about half have used the term “promising.”
“Today, patients and their families can easily find FDA press releases on the Internet, or they often hear about them in the news,” said study author Steven Woloshin, MD, of The Dartmouth Institute for Health Policy and Clinical Practice in Lebanon, New Hampshire.
“But the reality is that unless patients fully understand how the FDA is using the term ‘breakthrough,’ they may have unwarranted confidence in the evidence supporting drug claims. So we thought it was important to test how these terms affect the judgement of people without medical training.”
Survey details
The investigators conducted an online survey of 597 Americans. Participants were randomly given 1 of 5 short descriptions of a recently approved drug.
The descriptions were based on an FDA press release for a metastatic lung cancer breakthrough-designated drug that was conditionally approved based on the surrogate outcome of tumor shrinkage.
The first, “facts-only” description described the drug as meeting the criteria for breakthrough designation but did not actually use the term “breakthrough.”
A second and a third description included the facts and added the terms “breakthrough” and “promising,” respectively.
The fourth, “tentative” description included the facts, used the word “breakthrough,” and used the following FDA-required language for professional labeling:
The FDA pointed out that the drug was approved based on tumor shrinkage but that an improvement in survival or disease-related symptoms has not been established. Continued approval for this indication may be contingent upon verification and description of clinical benefit in confirmatory trials.
The fifth, “definitive” description included the same information as the tentative description, but “may be contingent” was changed to “is contingent.”
Participants were then asked to judge the drug’s benefit, harm, and strength of evidence.
Results
The investigators found that adding either “breakthrough” or “promising” in the description significantly increased the percentage of participants who rated the drug as “very” or “completely” effective compared with the facts-only description—23% and 25% vs 11%.
Adding “breakthrough” or “promising” to the description also significantly increased the number of people who reported believing that evidence supporting the drug is “strong” or “extremely strong”—59% and 63% vs 43%.
At the same time, adding either the tentative or definitive explanations significantly reduced the percentage of participants who believed (incorrectly) that the drug had been “proven to save lives”—16% tentative and 10% definitive vs 31% breakthrough.
Finally, when participants were asked which of 2 drugs—one described as “breakthrough,” the other as meeting the breakthrough criteria—they would take for a potentially deadly condition, 92% chose the “breakthrough” drug.
“Our findings clearly indicate that words like ‘breakthrough’ and ‘promising’ increase people’s beliefs in a drug’s effectiveness,” said Lisa Schwartz, MD, of The Dartmouth Institute for Health Policy and Clinical Practice.
“In light of [the findings], press releases with neutral terms and that clearly explain the limited evidence supporting what breakthrough designation and accelerated approval mean might help consumers make more accurate judgements about these drugs.” ![]()

Photo courtesy of the FDA
Using the words “breakthrough” and “promising” to describe new drugs affects the public’s perception of the drugs’ effectiveness, according to a study published in JAMA Internal Medicine.
Investigators noted that, in everyday usage, the term “breakthrough” represents a highly significant or definitive advance.
However, the US Food and Drug Administration’s (FDA’s) “breakthrough therapy designation” has a different meaning.
Since the Safety and Innovation Act became law in 2012, the FDA can assign breakthrough designation to a drug that “treats a serious or life-threatening condition” and “may demonstrate a substantial improvement . . . over available therapies” based on preliminary evidence.
And since the creation of the Safety and Innovation Act, all FDA press releases announcing the approval of breakthrough-designated drugs have used the term “breakthrough,” while about half have used the term “promising.”
“Today, patients and their families can easily find FDA press releases on the Internet, or they often hear about them in the news,” said study author Steven Woloshin, MD, of The Dartmouth Institute for Health Policy and Clinical Practice in Lebanon, New Hampshire.
“But the reality is that unless patients fully understand how the FDA is using the term ‘breakthrough,’ they may have unwarranted confidence in the evidence supporting drug claims. So we thought it was important to test how these terms affect the judgement of people without medical training.”
Survey details
The investigators conducted an online survey of 597 Americans. Participants were randomly given 1 of 5 short descriptions of a recently approved drug.
The descriptions were based on an FDA press release for a metastatic lung cancer breakthrough-designated drug that was conditionally approved based on the surrogate outcome of tumor shrinkage.
The first, “facts-only” description described the drug as meeting the criteria for breakthrough designation but did not actually use the term “breakthrough.”
A second and a third description included the facts and added the terms “breakthrough” and “promising,” respectively.
The fourth, “tentative” description included the facts, used the word “breakthrough,” and used the following FDA-required language for professional labeling:
The FDA pointed out that the drug was approved based on tumor shrinkage but that an improvement in survival or disease-related symptoms has not been established. Continued approval for this indication may be contingent upon verification and description of clinical benefit in confirmatory trials.
The fifth, “definitive” description included the same information as the tentative description, but “may be contingent” was changed to “is contingent.”
Participants were then asked to judge the drug’s benefit, harm, and strength of evidence.
Results
The investigators found that adding either “breakthrough” or “promising” in the description significantly increased the percentage of participants who rated the drug as “very” or “completely” effective compared with the facts-only description—23% and 25% vs 11%.
Adding “breakthrough” or “promising” to the description also significantly increased the number of people who reported believing that evidence supporting the drug is “strong” or “extremely strong”—59% and 63% vs 43%.
At the same time, adding either the tentative or definitive explanations significantly reduced the percentage of participants who believed (incorrectly) that the drug had been “proven to save lives”—16% tentative and 10% definitive vs 31% breakthrough.
Finally, when participants were asked which of 2 drugs—one described as “breakthrough,” the other as meeting the breakthrough criteria—they would take for a potentially deadly condition, 92% chose the “breakthrough” drug.
“Our findings clearly indicate that words like ‘breakthrough’ and ‘promising’ increase people’s beliefs in a drug’s effectiveness,” said Lisa Schwartz, MD, of The Dartmouth Institute for Health Policy and Clinical Practice.
“In light of [the findings], press releases with neutral terms and that clearly explain the limited evidence supporting what breakthrough designation and accelerated approval mean might help consumers make more accurate judgements about these drugs.” ![]()
Creating off-the-shelf VSTs

Photo courtesy of Baylor
College of Medicine
NEW YORK—Researchers are creating virus-specific T cells (VST) to treat and prevent viral infections in patients who undergo hematopoietic stem cell transplant.
Thus far, the group has modified T cells with 5 viral vectors—Epstein-Barr virus (EBV), cytomegalovirus (CMV), adenovirus (ADV), BK virus (BKV), and human herpesvirus 6 (HHV6)—and are devising methods whereby these VSTs can be made readily available, off-the-shelf products.
Helen Heslop, MD, of Baylor College of Medicine in Houston, Texas, described the efforts of the Baylor research team at the inaugural CRI-CIMT-EATI-AACR International Cancer Immunotherapy Conference.
The team’s tri-virus and 5-virus VST approaches have been described previously. Here, we focus on the team’s efforts to create an off-the-shelf product.
A companion story describes the team’s VST approach to treating type 2 EBV-associated lymphomas.
Despite promising results with these earlier methodologies both as prophylaxis and treatment after stem cell transplant, problems existed with the approaches that would impede broader implementation of VSTs.
“Most of these initial methodologies were complex,” Dr Heslop said.
And even though the production time of the 5-virus VSTs is only 10 days, it does not allow for urgent use.
To solve this problem, the researchers are developing VSTs as off-the-shelf, third-party banked cells for patients who don’t have the time to wait for donor-specific cells to be made.
“The strategy here is that we make lines that are well-characterized but are HLA-restricted and tied to specific viruses,” Dr Heslop said.
The cells are then cryopreserved so that they’re available, and patients receive VSTs according to their HLA type and the line that has suitable activity for their infections.
“We initially evaluated this approach through a multicenter study sponsored by the NHLBI [National Heart, Lung, and Blood Institute],” Dr Heslop said.
Tri-virus approach
The researchers used the original tri-virus methodology, which was lengthy, “but, in this case, because the cells were already available, the time was not a major issue,” Dr Heslop said.
The team also made some new lines for donors with common alleles. In all, they had 32 lines available while the study was running.
They treated 50 patients: 23 received VSTs for CMV, 18 for ADV, and 9 for EBV.
One patient who had CMV colitis received the VSTs and had a complete response (CR), as evidenced by a normal endoscopy with no viral inclusions.
Another patient had EBV persistent after 6 doses of rituximab. One month after receiving the VST infusion, the patient had a CR, even though the cell line had only 1 class 2 allele. And the patient has sustained the CR for several years.
The overall response rate for the study is 74.0% at day 42 after infusion.
The response rate was not significantly different for each virus, Dr Heslop pointed out. Patients with CMV had a 73.9% response rate, those with EBV had a 66.7% response rate, and those with ADV had a 77.8% response rate.
“This is a little bit lower than with the donor-specific T cells, but I think it’s still a promising approach,” she added.
5-virus peptide mix
The researchers are now evaluating the more rapid, 5-virus peptide mix method for creating off-the-shelf VSTs.
This method replaced live viruses with overlapping peptide pools and added immunogenic antigens for 5 viruses, including BKV and HHV6. The process takes only 10 days to produce VSTs.
The team has identified a line for over 90% of the patients screened.
“That’s because we will accept a line that’s only matched at 1 HLA allele, as long as we have activity against the infecting virus through the shared allele,” Dr Heslop explained.
So they’re conducting a clinical trial using this method, and thus far, they have enrolled 22 patients. Sixteen patients have had 1 infusion, and 6 patients have had multiple infusions.
The 22 patients had 25 infections: 10 CMV, 10 BKV, 2 EBV, 2 ADV, and 1 HHV6.
The overall response rate is 88%. Nine of 10 responses in patients with BKV were partial because BK is difficult to clear from urine. However, the BK patients who had hemorrhagic cystitis all had symptomatic improvement.
Dr Heslop pointed out that these results are similar to those at other centers using EBV third-party T cells.
The initial third-party studies were done in Scotland and had a response rate of about 60%, which increased to around 80% in follow-up studies, where they characterized the T cells more extensively.
Both donor-specific and third-party VSTs have low toxicity, sustained response rates, and activity confirmed by studies in multiple centers.
Dr Heslop believes the rapid manufacturing methodologies will facilitate definitive clinical trials.
She said Cell Medica provided support for some of the trials with EBV tumors. ![]()

Photo courtesy of Baylor
College of Medicine
NEW YORK—Researchers are creating virus-specific T cells (VST) to treat and prevent viral infections in patients who undergo hematopoietic stem cell transplant.
Thus far, the group has modified T cells with 5 viral vectors—Epstein-Barr virus (EBV), cytomegalovirus (CMV), adenovirus (ADV), BK virus (BKV), and human herpesvirus 6 (HHV6)—and are devising methods whereby these VSTs can be made readily available, off-the-shelf products.
Helen Heslop, MD, of Baylor College of Medicine in Houston, Texas, described the efforts of the Baylor research team at the inaugural CRI-CIMT-EATI-AACR International Cancer Immunotherapy Conference.
The team’s tri-virus and 5-virus VST approaches have been described previously. Here, we focus on the team’s efforts to create an off-the-shelf product.
A companion story describes the team’s VST approach to treating type 2 EBV-associated lymphomas.
Despite promising results with these earlier methodologies both as prophylaxis and treatment after stem cell transplant, problems existed with the approaches that would impede broader implementation of VSTs.
“Most of these initial methodologies were complex,” Dr Heslop said.
And even though the production time of the 5-virus VSTs is only 10 days, it does not allow for urgent use.
To solve this problem, the researchers are developing VSTs as off-the-shelf, third-party banked cells for patients who don’t have the time to wait for donor-specific cells to be made.
“The strategy here is that we make lines that are well-characterized but are HLA-restricted and tied to specific viruses,” Dr Heslop said.
The cells are then cryopreserved so that they’re available, and patients receive VSTs according to their HLA type and the line that has suitable activity for their infections.
“We initially evaluated this approach through a multicenter study sponsored by the NHLBI [National Heart, Lung, and Blood Institute],” Dr Heslop said.
Tri-virus approach
The researchers used the original tri-virus methodology, which was lengthy, “but, in this case, because the cells were already available, the time was not a major issue,” Dr Heslop said.
The team also made some new lines for donors with common alleles. In all, they had 32 lines available while the study was running.
They treated 50 patients: 23 received VSTs for CMV, 18 for ADV, and 9 for EBV.
One patient who had CMV colitis received the VSTs and had a complete response (CR), as evidenced by a normal endoscopy with no viral inclusions.
Another patient had EBV persistent after 6 doses of rituximab. One month after receiving the VST infusion, the patient had a CR, even though the cell line had only 1 class 2 allele. And the patient has sustained the CR for several years.
The overall response rate for the study is 74.0% at day 42 after infusion.
The response rate was not significantly different for each virus, Dr Heslop pointed out. Patients with CMV had a 73.9% response rate, those with EBV had a 66.7% response rate, and those with ADV had a 77.8% response rate.
“This is a little bit lower than with the donor-specific T cells, but I think it’s still a promising approach,” she added.
5-virus peptide mix
The researchers are now evaluating the more rapid, 5-virus peptide mix method for creating off-the-shelf VSTs.
This method replaced live viruses with overlapping peptide pools and added immunogenic antigens for 5 viruses, including BKV and HHV6. The process takes only 10 days to produce VSTs.
The team has identified a line for over 90% of the patients screened.
“That’s because we will accept a line that’s only matched at 1 HLA allele, as long as we have activity against the infecting virus through the shared allele,” Dr Heslop explained.
So they’re conducting a clinical trial using this method, and thus far, they have enrolled 22 patients. Sixteen patients have had 1 infusion, and 6 patients have had multiple infusions.
The 22 patients had 25 infections: 10 CMV, 10 BKV, 2 EBV, 2 ADV, and 1 HHV6.
The overall response rate is 88%. Nine of 10 responses in patients with BKV were partial because BK is difficult to clear from urine. However, the BK patients who had hemorrhagic cystitis all had symptomatic improvement.
Dr Heslop pointed out that these results are similar to those at other centers using EBV third-party T cells.
The initial third-party studies were done in Scotland and had a response rate of about 60%, which increased to around 80% in follow-up studies, where they characterized the T cells more extensively.
Both donor-specific and third-party VSTs have low toxicity, sustained response rates, and activity confirmed by studies in multiple centers.
Dr Heslop believes the rapid manufacturing methodologies will facilitate definitive clinical trials.
She said Cell Medica provided support for some of the trials with EBV tumors. ![]()

Photo courtesy of Baylor
College of Medicine
NEW YORK—Researchers are creating virus-specific T cells (VST) to treat and prevent viral infections in patients who undergo hematopoietic stem cell transplant.
Thus far, the group has modified T cells with 5 viral vectors—Epstein-Barr virus (EBV), cytomegalovirus (CMV), adenovirus (ADV), BK virus (BKV), and human herpesvirus 6 (HHV6)—and are devising methods whereby these VSTs can be made readily available, off-the-shelf products.
Helen Heslop, MD, of Baylor College of Medicine in Houston, Texas, described the efforts of the Baylor research team at the inaugural CRI-CIMT-EATI-AACR International Cancer Immunotherapy Conference.
The team’s tri-virus and 5-virus VST approaches have been described previously. Here, we focus on the team’s efforts to create an off-the-shelf product.
A companion story describes the team’s VST approach to treating type 2 EBV-associated lymphomas.
Despite promising results with these earlier methodologies both as prophylaxis and treatment after stem cell transplant, problems existed with the approaches that would impede broader implementation of VSTs.
“Most of these initial methodologies were complex,” Dr Heslop said.
And even though the production time of the 5-virus VSTs is only 10 days, it does not allow for urgent use.
To solve this problem, the researchers are developing VSTs as off-the-shelf, third-party banked cells for patients who don’t have the time to wait for donor-specific cells to be made.
“The strategy here is that we make lines that are well-characterized but are HLA-restricted and tied to specific viruses,” Dr Heslop said.
The cells are then cryopreserved so that they’re available, and patients receive VSTs according to their HLA type and the line that has suitable activity for their infections.
“We initially evaluated this approach through a multicenter study sponsored by the NHLBI [National Heart, Lung, and Blood Institute],” Dr Heslop said.
Tri-virus approach
The researchers used the original tri-virus methodology, which was lengthy, “but, in this case, because the cells were already available, the time was not a major issue,” Dr Heslop said.
The team also made some new lines for donors with common alleles. In all, they had 32 lines available while the study was running.
They treated 50 patients: 23 received VSTs for CMV, 18 for ADV, and 9 for EBV.
One patient who had CMV colitis received the VSTs and had a complete response (CR), as evidenced by a normal endoscopy with no viral inclusions.
Another patient had EBV persistent after 6 doses of rituximab. One month after receiving the VST infusion, the patient had a CR, even though the cell line had only 1 class 2 allele. And the patient has sustained the CR for several years.
The overall response rate for the study is 74.0% at day 42 after infusion.
The response rate was not significantly different for each virus, Dr Heslop pointed out. Patients with CMV had a 73.9% response rate, those with EBV had a 66.7% response rate, and those with ADV had a 77.8% response rate.
“This is a little bit lower than with the donor-specific T cells, but I think it’s still a promising approach,” she added.
5-virus peptide mix
The researchers are now evaluating the more rapid, 5-virus peptide mix method for creating off-the-shelf VSTs.
This method replaced live viruses with overlapping peptide pools and added immunogenic antigens for 5 viruses, including BKV and HHV6. The process takes only 10 days to produce VSTs.
The team has identified a line for over 90% of the patients screened.
“That’s because we will accept a line that’s only matched at 1 HLA allele, as long as we have activity against the infecting virus through the shared allele,” Dr Heslop explained.
So they’re conducting a clinical trial using this method, and thus far, they have enrolled 22 patients. Sixteen patients have had 1 infusion, and 6 patients have had multiple infusions.
The 22 patients had 25 infections: 10 CMV, 10 BKV, 2 EBV, 2 ADV, and 1 HHV6.
The overall response rate is 88%. Nine of 10 responses in patients with BKV were partial because BK is difficult to clear from urine. However, the BK patients who had hemorrhagic cystitis all had symptomatic improvement.
Dr Heslop pointed out that these results are similar to those at other centers using EBV third-party T cells.
The initial third-party studies were done in Scotland and had a response rate of about 60%, which increased to around 80% in follow-up studies, where they characterized the T cells more extensively.
Both donor-specific and third-party VSTs have low toxicity, sustained response rates, and activity confirmed by studies in multiple centers.
Dr Heslop believes the rapid manufacturing methodologies will facilitate definitive clinical trials.
She said Cell Medica provided support for some of the trials with EBV tumors.
Trio wins Nobel Prize for parasite-related discoveries

Three researchers have won the 2015 Nobel Prize in Physiology or Medicine for discoveries related to parasitic diseases.
One half of the prize was awarded to Youyou Tu for discoveries concerning a novel therapy against malaria—artemisinin.
The other half of the prize was awarded to William C. Campbell, PhD, and Satoshi Ōmura, PhD, for their discoveries concerning a novel therapy against infections caused by roundworm parasites.
Drs Ōmura and Campbell discovered the drug avermectin. A derivative of this drug has lowered the incidence of river blindness and lymphatic filariasis and demonstrated efficacy against other parasitic diseases.
Artemisinin
Before artemisinin came into use, malaria was treated with chloroquine or quinine—with declining success. By the late 1960s, efforts to eradicate malaria had failed, and the disease was on the rise.
At that time, Tu turned to traditional herbal medicine to tackle the challenge of developing novel malaria therapies. From a large-scale screen of herbal remedies in malaria-infected animals, an extract from the plant Artemisia annua emerged as an interesting candidate.
However, the results were inconsistent. So Tu revisited the ancient literature and discovered clues that guided her in her quest to extract the active component from Artemisia annua. Tu was the first to show that this component, later called artemisinin, was effective against the malaria parasite in animals and humans.
Artemisinin is now used in all malaria-ridden parts of the world. When used in combination therapy, it is estimated to reduce mortality from malaria by more than 20% overall and by more than 30% in children.
Avermectin
The discovery of avermectin began with Streptomyces, bacteria that live in the soil and are known to produce agents with antibacterial activities.
Dr Ōmura isolated new strains of Streptomyces from soil samples and cultured them in the lab. He selected about 50 of the most promising cultures to analyze for their activity against harmful microorganisms. One of these cultures turned out to be Streptomyces avermitilis, the source of avermectin.
Dr Campbell acquired Dr Ōmura’s Streptomyces cultures and explored their efficacy. Dr Campbell showed that a component from one of the cultures could combat parasites in domestic and farm animals.
The bioactive agent was purified and named avermectin. It was subsequently modified to a more effective compound called ivermectin. Ivermectin turned out to be effective against a variety of parasites, including those that cause river blindness and lymphatic filariasis.
Today, ivermectin is used in all parts of the world that are plagued by parasitic diseases. The drug has proven effective against a range of parasites and has limited side effects. Thanks to ivermectin, river blindness and lymphatic filariasis are on the verge of eradication.
About the winners
Youyou Tu was born in 1930 in China. She graduated from Beijing Medical University in 1955. Tu has worked at the China Academy of Traditional Chinese Medicine since 1965. She has been chief professor there since 2000.
William C. Campbell was born in 1930 in Ramelton, Ireland. He received a BA from Trinity College, University of Dublin, in Ireland in 1952. He received a PhD from the University of Wisconsin in Madison, Wisconsin, in 1957.
From 1957 to 1990, Dr Campbell was with the Merck Institute for Therapeutic Research, from 1984 to 1990 as a senior scientist and director for assay research and development. Dr Campbell is currently a research fellow emeritus at Drew University in Madison, New Jersey.
Satoshi Ōmura was born in 1935 in the Yamanashi Prefecture, Japan. He received a PhD in pharmaceutical sciences in 1968 from the University of Tokyo and a PhD in chemistry in 1970 from Tokyo University of Science.
Dr Ōmura was a researcher at the Kitasato Institute in Japan from 1965 to 1971 and a professor at Kitasato University from 1975 to 2007. Since 2007, Dr Ōmura has been a professor emeritus at Kitasato University.

Three researchers have won the 2015 Nobel Prize in Physiology or Medicine for discoveries related to parasitic diseases.
One half of the prize was awarded to Youyou Tu for discoveries concerning a novel therapy against malaria—artemisinin.
The other half of the prize was awarded to William C. Campbell, PhD, and Satoshi Ōmura, PhD, for their discoveries concerning a novel therapy against infections caused by roundworm parasites.
Drs Ōmura and Campbell discovered the drug avermectin. A derivative of this drug has lowered the incidence of river blindness and lymphatic filariasis and demonstrated efficacy against other parasitic diseases.
Artemisinin
Before artemisinin came into use, malaria was treated with chloroquine or quinine—with declining success. By the late 1960s, efforts to eradicate malaria had failed, and the disease was on the rise.
At that time, Tu turned to traditional herbal medicine to tackle the challenge of developing novel malaria therapies. From a large-scale screen of herbal remedies in malaria-infected animals, an extract from the plant Artemisia annua emerged as an interesting candidate.
However, the results were inconsistent. So Tu revisited the ancient literature and discovered clues that guided her in her quest to extract the active component from Artemisia annua. Tu was the first to show that this component, later called artemisinin, was effective against the malaria parasite in animals and humans.
Artemisinin is now used in all malaria-ridden parts of the world. When used in combination therapy, it is estimated to reduce mortality from malaria by more than 20% overall and by more than 30% in children.
Avermectin
The discovery of avermectin began with Streptomyces, bacteria that live in the soil and are known to produce agents with antibacterial activities.
Dr Ōmura isolated new strains of Streptomyces from soil samples and cultured them in the lab. He selected about 50 of the most promising cultures to analyze for their activity against harmful microorganisms. One of these cultures turned out to be Streptomyces avermitilis, the source of avermectin.
Dr Campbell acquired Dr Ōmura’s Streptomyces cultures and explored their efficacy. Dr Campbell showed that a component from one of the cultures could combat parasites in domestic and farm animals.
The bioactive agent was purified and named avermectin. It was subsequently modified to a more effective compound called ivermectin. Ivermectin turned out to be effective against a variety of parasites, including those that cause river blindness and lymphatic filariasis.
Today, ivermectin is used in all parts of the world that are plagued by parasitic diseases. The drug has proven effective against a range of parasites and has limited side effects. Thanks to ivermectin, river blindness and lymphatic filariasis are on the verge of eradication.
About the winners
Youyou Tu was born in 1930 in China. She graduated from Beijing Medical University in 1955. Tu has worked at the China Academy of Traditional Chinese Medicine since 1965. She has been chief professor there since 2000.
William C. Campbell was born in 1930 in Ramelton, Ireland. He received a BA from Trinity College, University of Dublin, in Ireland in 1952. He received a PhD from the University of Wisconsin in Madison, Wisconsin, in 1957.
From 1957 to 1990, Dr Campbell was with the Merck Institute for Therapeutic Research, from 1984 to 1990 as a senior scientist and director for assay research and development. Dr Campbell is currently a research fellow emeritus at Drew University in Madison, New Jersey.
Satoshi Ōmura was born in 1935 in the Yamanashi Prefecture, Japan. He received a PhD in pharmaceutical sciences in 1968 from the University of Tokyo and a PhD in chemistry in 1970 from Tokyo University of Science.
Dr Ōmura was a researcher at the Kitasato Institute in Japan from 1965 to 1971 and a professor at Kitasato University from 1975 to 2007. Since 2007, Dr Ōmura has been a professor emeritus at Kitasato University.

Three researchers have won the 2015 Nobel Prize in Physiology or Medicine for discoveries related to parasitic diseases.
One half of the prize was awarded to Youyou Tu for discoveries concerning a novel therapy against malaria—artemisinin.
The other half of the prize was awarded to William C. Campbell, PhD, and Satoshi Ōmura, PhD, for their discoveries concerning a novel therapy against infections caused by roundworm parasites.
Drs Ōmura and Campbell discovered the drug avermectin. A derivative of this drug has lowered the incidence of river blindness and lymphatic filariasis and demonstrated efficacy against other parasitic diseases.
Artemisinin
Before artemisinin came into use, malaria was treated with chloroquine or quinine—with declining success. By the late 1960s, efforts to eradicate malaria had failed, and the disease was on the rise.
At that time, Tu turned to traditional herbal medicine to tackle the challenge of developing novel malaria therapies. From a large-scale screen of herbal remedies in malaria-infected animals, an extract from the plant Artemisia annua emerged as an interesting candidate.
However, the results were inconsistent. So Tu revisited the ancient literature and discovered clues that guided her in her quest to extract the active component from Artemisia annua. Tu was the first to show that this component, later called artemisinin, was effective against the malaria parasite in animals and humans.
Artemisinin is now used in all malaria-ridden parts of the world. When used in combination therapy, it is estimated to reduce mortality from malaria by more than 20% overall and by more than 30% in children.
Avermectin
The discovery of avermectin began with Streptomyces, bacteria that live in the soil and are known to produce agents with antibacterial activities.
Dr Ōmura isolated new strains of Streptomyces from soil samples and cultured them in the lab. He selected about 50 of the most promising cultures to analyze for their activity against harmful microorganisms. One of these cultures turned out to be Streptomyces avermitilis, the source of avermectin.
Dr Campbell acquired Dr Ōmura’s Streptomyces cultures and explored their efficacy. Dr Campbell showed that a component from one of the cultures could combat parasites in domestic and farm animals.
The bioactive agent was purified and named avermectin. It was subsequently modified to a more effective compound called ivermectin. Ivermectin turned out to be effective against a variety of parasites, including those that cause river blindness and lymphatic filariasis.
Today, ivermectin is used in all parts of the world that are plagued by parasitic diseases. The drug has proven effective against a range of parasites and has limited side effects. Thanks to ivermectin, river blindness and lymphatic filariasis are on the verge of eradication.
About the winners
Youyou Tu was born in 1930 in China. She graduated from Beijing Medical University in 1955. Tu has worked at the China Academy of Traditional Chinese Medicine since 1965. She has been chief professor there since 2000.
William C. Campbell was born in 1930 in Ramelton, Ireland. He received a BA from Trinity College, University of Dublin, in Ireland in 1952. He received a PhD from the University of Wisconsin in Madison, Wisconsin, in 1957.
From 1957 to 1990, Dr Campbell was with the Merck Institute for Therapeutic Research, from 1984 to 1990 as a senior scientist and director for assay research and development. Dr Campbell is currently a research fellow emeritus at Drew University in Madison, New Jersey.
Satoshi Ōmura was born in 1935 in the Yamanashi Prefecture, Japan. He received a PhD in pharmaceutical sciences in 1968 from the University of Tokyo and a PhD in chemistry in 1970 from Tokyo University of Science.
Dr Ōmura was a researcher at the Kitasato Institute in Japan from 1965 to 1971 and a professor at Kitasato University from 1975 to 2007. Since 2007, Dr Ōmura has been a professor emeritus at Kitasato University.
NICE can’t recommend new sepsis tests
Photo by William Weinert
The UK’s National Institute for Health and Care Excellence (NICE) has said there is not enough evidence to recommend 3 new blood tests for routine use in the National Health Service.
The tests are designed to identify bacteria and fungi in the bloodstream more rapidly than current tests.
According to NICE, there is too much uncertainty regarding the accuracy of the new tests and the size of benefit they might confer for patients with suspected sepsis.
The tests in question are LightCycler SeptiFast Test MGRADE (Roche Diagnostics), SepsiTest (Molzym Molecular Diagnostics), and IRIDICA BAC BSI assay (Abbott Diagnostics).
They are used to analyze whole blood samples for bacterial and fungal DNA, which may identify pathogens earlier than microbiology techniques. Microbiology techniques require blood samples to be incubated and cultured before pathogens can be identified.
The new tests are designed to enable earlier targeted treatment for patients with sepsis and reduce the use of broad-spectrum antimicrobials, which could help reduce future antimicrobial resistance.
“Rapid molecular tests that can identify which pathogens are the cause of an infection in hours rather than the days typically needed for traditional microbiology tests could ensure the right antibiotics are used much earlier in treatment,” said Carole Longson, NICE Health Technology Evaluation Centre Director.
“This, in turn, could improve outcomes for patients with suspected sepsis as well as help to reduce the spread of resistant microbes. However, the committee [advising NICE] concluded that the tests may offer clinical benefit, but there is too much uncertainty in the size of the benefit to determine the effect of introducing the tests into clinical practice.”
“The committee also concluded that, although the rapid molecular tests might provide results more quickly, there was too much uncertainty in the accuracy of the tests for clinicians to be able to base a decision on whether to withdraw or continue antibiotics. The committee therefore decided that further research should be encouraged to determine the clinical scenarios in which the tests may offer most benefit.”
NICE’s draft diagnostics guidance on the LightCycler SeptiFast Test MGRADE, SepsiTest, and IRIDICA BAC BSI assay is available on the NICE website. The closing date for comments on this draft guidance is October 21, 2015.

Photo by William Weinert
The UK’s National Institute for Health and Care Excellence (NICE) has said there is not enough evidence to recommend 3 new blood tests for routine use in the National Health Service.
The tests are designed to identify bacteria and fungi in the bloodstream more rapidly than current tests.
According to NICE, there is too much uncertainty regarding the accuracy of the new tests and the size of benefit they might confer for patients with suspected sepsis.
The tests in question are LightCycler SeptiFast Test MGRADE (Roche Diagnostics), SepsiTest (Molzym Molecular Diagnostics), and IRIDICA BAC BSI assay (Abbott Diagnostics).
They are used to analyze whole blood samples for bacterial and fungal DNA, which may identify pathogens earlier than microbiology techniques. Microbiology techniques require blood samples to be incubated and cultured before pathogens can be identified.
The new tests are designed to enable earlier targeted treatment for patients with sepsis and reduce the use of broad-spectrum antimicrobials, which could help reduce future antimicrobial resistance.
“Rapid molecular tests that can identify which pathogens are the cause of an infection in hours rather than the days typically needed for traditional microbiology tests could ensure the right antibiotics are used much earlier in treatment,” said Carole Longson, NICE Health Technology Evaluation Centre Director.
“This, in turn, could improve outcomes for patients with suspected sepsis as well as help to reduce the spread of resistant microbes. However, the committee [advising NICE] concluded that the tests may offer clinical benefit, but there is too much uncertainty in the size of the benefit to determine the effect of introducing the tests into clinical practice.”
“The committee also concluded that, although the rapid molecular tests might provide results more quickly, there was too much uncertainty in the accuracy of the tests for clinicians to be able to base a decision on whether to withdraw or continue antibiotics. The committee therefore decided that further research should be encouraged to determine the clinical scenarios in which the tests may offer most benefit.”
NICE’s draft diagnostics guidance on the LightCycler SeptiFast Test MGRADE, SepsiTest, and IRIDICA BAC BSI assay is available on the NICE website. The closing date for comments on this draft guidance is October 21, 2015.

Photo by William Weinert
The UK’s National Institute for Health and Care Excellence (NICE) has said there is not enough evidence to recommend 3 new blood tests for routine use in the National Health Service.
The tests are designed to identify bacteria and fungi in the bloodstream more rapidly than current tests.
According to NICE, there is too much uncertainty regarding the accuracy of the new tests and the size of benefit they might confer for patients with suspected sepsis.
The tests in question are LightCycler SeptiFast Test MGRADE (Roche Diagnostics), SepsiTest (Molzym Molecular Diagnostics), and IRIDICA BAC BSI assay (Abbott Diagnostics).
They are used to analyze whole blood samples for bacterial and fungal DNA, which may identify pathogens earlier than microbiology techniques. Microbiology techniques require blood samples to be incubated and cultured before pathogens can be identified.
The new tests are designed to enable earlier targeted treatment for patients with sepsis and reduce the use of broad-spectrum antimicrobials, which could help reduce future antimicrobial resistance.
“Rapid molecular tests that can identify which pathogens are the cause of an infection in hours rather than the days typically needed for traditional microbiology tests could ensure the right antibiotics are used much earlier in treatment,” said Carole Longson, NICE Health Technology Evaluation Centre Director.
“This, in turn, could improve outcomes for patients with suspected sepsis as well as help to reduce the spread of resistant microbes. However, the committee [advising NICE] concluded that the tests may offer clinical benefit, but there is too much uncertainty in the size of the benefit to determine the effect of introducing the tests into clinical practice.”
“The committee also concluded that, although the rapid molecular tests might provide results more quickly, there was too much uncertainty in the accuracy of the tests for clinicians to be able to base a decision on whether to withdraw or continue antibiotics. The committee therefore decided that further research should be encouraged to determine the clinical scenarios in which the tests may offer most benefit.”
NICE’s draft diagnostics guidance on the LightCycler SeptiFast Test MGRADE, SepsiTest, and IRIDICA BAC BSI assay is available on the NICE website. The closing date for comments on this draft guidance is October 21, 2015.
Group identifies malaria resistance locus

outside of Nairobi, Kenya
Photo by Gabrielle Tenenbaum
Researchers say they have identified genetic variants that protect African children from developing severe malaria, in some cases nearly halving a child’s chance of developing the disease.
The variants are at a locus located next to a cluster of genes that are responsible for creating the receptors the malaria parasite Plasmodium falciparum uses to infect red blood cells.
The researchers described their findings in a letter to Nature.
“The risk of developing severe malaria turns out to be strongly linked to the process by which the malaria parasite gains entry to the human red blood cell,” said Dr Kevin Marsh, of the Kemri-Wellcome Research Programme in Kilifi, Kenya.
“This study strengthens the argument for focusing on the malaria side of the parasite-human interaction in our search for new vaccine candidates.”
For this study, Dr Marsh and his colleagues analyzed data from 8 different African countries: Burkina Faso, Cameroon, Ghana, Kenya, Malawi, Mali, The Gambia, and Tanzania.
They compared the DNA of 5633 children with severe malaria and the DNA of 5919 children without severe malaria. The researchers then replicated their key findings in a further 14,000 children.
The locus the team identified is near a cluster of genes that code for glycophorins, which are involved in P falciparum’s invasion of red blood cells.
The researchers also found an allele that was common among children in Kenya. Having this allele reduced the risk of severe malaria by about 40% in Kenyan children, with a slightly smaller effect across all the other populations studied.
The team said this difference between populations could be due to the genetic features of the local malaria parasite in East Africa.
Balancing selection
The newly identified malaria resistance locus lies within a region of the genome where humans and chimpanzees have been known to share particular combinations of haplotypes.
This indicates that some of the variation seen in contemporary humans has been present for millions of years. The finding also suggests that this region of the genome is the subject of balancing selection.
Balancing selection happens when a particular genetic variant evolves because it confers health benefits, but it is carried by only a proportion of the population because it also has damaging consequences.
“These findings indicate that balancing selection and resistance to malaria are deeply intertwined themes in our ancient evolutionary history,” said Dr Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“This new resistance locus is particularly interesting because it lies so close to genes that are gatekeepers for the malaria parasite’s invasion machinery. We now need to drill down at this locus to characterize these complex patterns of genetic variation more precisely and to understand the molecular mechanisms by which they act.”

outside of Nairobi, Kenya
Photo by Gabrielle Tenenbaum
Researchers say they have identified genetic variants that protect African children from developing severe malaria, in some cases nearly halving a child’s chance of developing the disease.
The variants are at a locus located next to a cluster of genes that are responsible for creating the receptors the malaria parasite Plasmodium falciparum uses to infect red blood cells.
The researchers described their findings in a letter to Nature.
“The risk of developing severe malaria turns out to be strongly linked to the process by which the malaria parasite gains entry to the human red blood cell,” said Dr Kevin Marsh, of the Kemri-Wellcome Research Programme in Kilifi, Kenya.
“This study strengthens the argument for focusing on the malaria side of the parasite-human interaction in our search for new vaccine candidates.”
For this study, Dr Marsh and his colleagues analyzed data from 8 different African countries: Burkina Faso, Cameroon, Ghana, Kenya, Malawi, Mali, The Gambia, and Tanzania.
They compared the DNA of 5633 children with severe malaria and the DNA of 5919 children without severe malaria. The researchers then replicated their key findings in a further 14,000 children.
The locus the team identified is near a cluster of genes that code for glycophorins, which are involved in P falciparum’s invasion of red blood cells.
The researchers also found an allele that was common among children in Kenya. Having this allele reduced the risk of severe malaria by about 40% in Kenyan children, with a slightly smaller effect across all the other populations studied.
The team said this difference between populations could be due to the genetic features of the local malaria parasite in East Africa.
Balancing selection
The newly identified malaria resistance locus lies within a region of the genome where humans and chimpanzees have been known to share particular combinations of haplotypes.
This indicates that some of the variation seen in contemporary humans has been present for millions of years. The finding also suggests that this region of the genome is the subject of balancing selection.
Balancing selection happens when a particular genetic variant evolves because it confers health benefits, but it is carried by only a proportion of the population because it also has damaging consequences.
“These findings indicate that balancing selection and resistance to malaria are deeply intertwined themes in our ancient evolutionary history,” said Dr Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“This new resistance locus is particularly interesting because it lies so close to genes that are gatekeepers for the malaria parasite’s invasion machinery. We now need to drill down at this locus to characterize these complex patterns of genetic variation more precisely and to understand the molecular mechanisms by which they act.”

outside of Nairobi, Kenya
Photo by Gabrielle Tenenbaum
Researchers say they have identified genetic variants that protect African children from developing severe malaria, in some cases nearly halving a child’s chance of developing the disease.
The variants are at a locus located next to a cluster of genes that are responsible for creating the receptors the malaria parasite Plasmodium falciparum uses to infect red blood cells.
The researchers described their findings in a letter to Nature.
“The risk of developing severe malaria turns out to be strongly linked to the process by which the malaria parasite gains entry to the human red blood cell,” said Dr Kevin Marsh, of the Kemri-Wellcome Research Programme in Kilifi, Kenya.
“This study strengthens the argument for focusing on the malaria side of the parasite-human interaction in our search for new vaccine candidates.”
For this study, Dr Marsh and his colleagues analyzed data from 8 different African countries: Burkina Faso, Cameroon, Ghana, Kenya, Malawi, Mali, The Gambia, and Tanzania.
They compared the DNA of 5633 children with severe malaria and the DNA of 5919 children without severe malaria. The researchers then replicated their key findings in a further 14,000 children.
The locus the team identified is near a cluster of genes that code for glycophorins, which are involved in P falciparum’s invasion of red blood cells.
The researchers also found an allele that was common among children in Kenya. Having this allele reduced the risk of severe malaria by about 40% in Kenyan children, with a slightly smaller effect across all the other populations studied.
The team said this difference between populations could be due to the genetic features of the local malaria parasite in East Africa.
Balancing selection
The newly identified malaria resistance locus lies within a region of the genome where humans and chimpanzees have been known to share particular combinations of haplotypes.
This indicates that some of the variation seen in contemporary humans has been present for millions of years. The finding also suggests that this region of the genome is the subject of balancing selection.
Balancing selection happens when a particular genetic variant evolves because it confers health benefits, but it is carried by only a proportion of the population because it also has damaging consequences.
“These findings indicate that balancing selection and resistance to malaria are deeply intertwined themes in our ancient evolutionary history,” said Dr Dominic Kwiatkowski, of the Wellcome Trust Sanger Institute in Cambridge, UK.
“This new resistance locus is particularly interesting because it lies so close to genes that are gatekeepers for the malaria parasite’s invasion machinery. We now need to drill down at this locus to characterize these complex patterns of genetic variation more precisely and to understand the molecular mechanisms by which they act.”