User login
Guidelines reduce blood draws in critically ill kids
for a child in the pediatric ICU
Photo courtesy of
Johns Hopkins Medicine
New research suggests clinical practice guidelines can reduce the number of potentially unnecessary blood culture draws in critically ill children without endangering doctors’ ability to diagnose and treat sepsis.
The guidelines consist of 2 documents—a screening checklist and a decision algorithm.
In a single-center study, clinicians consulted these documents when considering ordering blood cultures for patients in a pediatric intensive care unit (ICU).
The clinicians said there was an immediate reduction in unnecessary blood draws after they began using these guidelines, and they were able to sustain the reduction over time.
Aaron Milstone, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland, and his colleagues described these results in JAMA Pediatrics.
The guidelines were created by a team of nurses, vascular access specialists, and physicians across specialties. The team created a fever/sepsis screening checklist and an accompanying decision-making flow chart designed to guide clinicians in the decision to draw blood.
These tools were posted in the pediatric ICU at The Johns Hopkins Hospital with instructions to be completed at the bedside by nurses and physicians. Each week, the team would meet to evaluate the data gathered, review how many cultures were sent from the unit, and discuss in detail individual cases where blood draws were necessary.
The researchers compared patient length of stay, mortality, readmission, and the number of episodes of suspected septic shock at the hospital before and after this intervention was implemented.
In the year before the team introduced the tools, there were 2204 patient visits to the pediatric ICU and 1807 blood cultures drawn.
After the intervention, there were 984 blood cultures drawn for 2356 patient visits, almost halving the number of blood cultures per patient day.
Comparing the pre- and post-intervention periods, there was no statistical difference in the occurrence of septic shock, hospital mortality, or hospital readmission.
Dr Milstone said this means patients experienced no increased risk of a missed sepsis diagnosis because of the intervention.
He and his colleagues said the future directions of this research include further exploring the implications this intervention may have for antibiotic use as well as working to implement the tools in other ICUs. The tools are already being tried at Johns Hopkins All Children’s Hospital in Florida and in the pediatric ICU at the University of Virginia. ![]()

for a child in the pediatric ICU
Photo courtesy of
Johns Hopkins Medicine
New research suggests clinical practice guidelines can reduce the number of potentially unnecessary blood culture draws in critically ill children without endangering doctors’ ability to diagnose and treat sepsis.
The guidelines consist of 2 documents—a screening checklist and a decision algorithm.
In a single-center study, clinicians consulted these documents when considering ordering blood cultures for patients in a pediatric intensive care unit (ICU).
The clinicians said there was an immediate reduction in unnecessary blood draws after they began using these guidelines, and they were able to sustain the reduction over time.
Aaron Milstone, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland, and his colleagues described these results in JAMA Pediatrics.
The guidelines were created by a team of nurses, vascular access specialists, and physicians across specialties. The team created a fever/sepsis screening checklist and an accompanying decision-making flow chart designed to guide clinicians in the decision to draw blood.
These tools were posted in the pediatric ICU at The Johns Hopkins Hospital with instructions to be completed at the bedside by nurses and physicians. Each week, the team would meet to evaluate the data gathered, review how many cultures were sent from the unit, and discuss in detail individual cases where blood draws were necessary.
The researchers compared patient length of stay, mortality, readmission, and the number of episodes of suspected septic shock at the hospital before and after this intervention was implemented.
In the year before the team introduced the tools, there were 2204 patient visits to the pediatric ICU and 1807 blood cultures drawn.
After the intervention, there were 984 blood cultures drawn for 2356 patient visits, almost halving the number of blood cultures per patient day.
Comparing the pre- and post-intervention periods, there was no statistical difference in the occurrence of septic shock, hospital mortality, or hospital readmission.
Dr Milstone said this means patients experienced no increased risk of a missed sepsis diagnosis because of the intervention.
He and his colleagues said the future directions of this research include further exploring the implications this intervention may have for antibiotic use as well as working to implement the tools in other ICUs. The tools are already being tried at Johns Hopkins All Children’s Hospital in Florida and in the pediatric ICU at the University of Virginia. ![]()

for a child in the pediatric ICU
Photo courtesy of
Johns Hopkins Medicine
New research suggests clinical practice guidelines can reduce the number of potentially unnecessary blood culture draws in critically ill children without endangering doctors’ ability to diagnose and treat sepsis.
The guidelines consist of 2 documents—a screening checklist and a decision algorithm.
In a single-center study, clinicians consulted these documents when considering ordering blood cultures for patients in a pediatric intensive care unit (ICU).
The clinicians said there was an immediate reduction in unnecessary blood draws after they began using these guidelines, and they were able to sustain the reduction over time.
Aaron Milstone, MD, of the Johns Hopkins University School of Medicine in Baltimore, Maryland, and his colleagues described these results in JAMA Pediatrics.
The guidelines were created by a team of nurses, vascular access specialists, and physicians across specialties. The team created a fever/sepsis screening checklist and an accompanying decision-making flow chart designed to guide clinicians in the decision to draw blood.
These tools were posted in the pediatric ICU at The Johns Hopkins Hospital with instructions to be completed at the bedside by nurses and physicians. Each week, the team would meet to evaluate the data gathered, review how many cultures were sent from the unit, and discuss in detail individual cases where blood draws were necessary.
The researchers compared patient length of stay, mortality, readmission, and the number of episodes of suspected septic shock at the hospital before and after this intervention was implemented.
In the year before the team introduced the tools, there were 2204 patient visits to the pediatric ICU and 1807 blood cultures drawn.
After the intervention, there were 984 blood cultures drawn for 2356 patient visits, almost halving the number of blood cultures per patient day.
Comparing the pre- and post-intervention periods, there was no statistical difference in the occurrence of septic shock, hospital mortality, or hospital readmission.
Dr Milstone said this means patients experienced no increased risk of a missed sepsis diagnosis because of the intervention.
He and his colleagues said the future directions of this research include further exploring the implications this intervention may have for antibiotic use as well as working to implement the tools in other ICUs. The tools are already being tried at Johns Hopkins All Children’s Hospital in Florida and in the pediatric ICU at the University of Virginia. ![]()
P falciparum malaria existed 2000 years ago, team says

individual from Velia, Italy
Photo courtesy of Luca
Bandioli, Pigorini Museum
An analysis of 2000-year-old human remains from several regions across the Italian peninsula has confirmed the presence of Plasmodium falciparum malaria during the Roman Empire, according to researchers.
The team found mitochondrial genomic evidence of P falciparum malaria, coaxed from the teeth of bodies buried in 3 Italian cemeteries, dating back to the Imperial period.
The researchers said these finding provide a key reference point for when and where the malaria parasite existed in humans, as well as more information about the evolution of human disease.
The team reported these findings in Current Biology.
“There is extensive written evidence describing fevers that sound like malaria in ancient Greece and Rome, but the specific malaria species responsible is unknown,” said study author Stephanie Marciniak, PhD, of Pennsylvania State University in University Park.
“Our data confirm that the species was likely Plasmodium falciparum and that it affected people in different ecological and cultural environments. These results open up new questions to explore, particularly how widespread this parasite was and what burden it placed upon communities in Imperial Roman Italy.”
Dr Marciniak and her colleagues sampled teeth taken from 58 adults interred at 3 Imperial period Italian cemeteries: Isola Sacra, Velia, and Vagnari.
Located on the coast, Velia and Isola Sacra were known as important port cities and trading centers. Vagnari is located further inland and believed to be the burial site of laborers who would have worked on a Roman rural estate.
The researchers mined tiny DNA fragments from dental pulp. They were able to extract, purify, and enrich specifically for the Plasmodium species known to infect humans.
The team noted that usable DNA is challenging to extract because the parasites primarily dwell within the bloodstream and organs, which decompose and break down over time—in this instance, over the course of 2 millennia.
However, the researchers recovered more than half of the P falciparum mitochondrial genome from 2 individuals from Velia and Vagnari. ![]()

individual from Velia, Italy
Photo courtesy of Luca
Bandioli, Pigorini Museum
An analysis of 2000-year-old human remains from several regions across the Italian peninsula has confirmed the presence of Plasmodium falciparum malaria during the Roman Empire, according to researchers.
The team found mitochondrial genomic evidence of P falciparum malaria, coaxed from the teeth of bodies buried in 3 Italian cemeteries, dating back to the Imperial period.
The researchers said these finding provide a key reference point for when and where the malaria parasite existed in humans, as well as more information about the evolution of human disease.
The team reported these findings in Current Biology.
“There is extensive written evidence describing fevers that sound like malaria in ancient Greece and Rome, but the specific malaria species responsible is unknown,” said study author Stephanie Marciniak, PhD, of Pennsylvania State University in University Park.
“Our data confirm that the species was likely Plasmodium falciparum and that it affected people in different ecological and cultural environments. These results open up new questions to explore, particularly how widespread this parasite was and what burden it placed upon communities in Imperial Roman Italy.”
Dr Marciniak and her colleagues sampled teeth taken from 58 adults interred at 3 Imperial period Italian cemeteries: Isola Sacra, Velia, and Vagnari.
Located on the coast, Velia and Isola Sacra were known as important port cities and trading centers. Vagnari is located further inland and believed to be the burial site of laborers who would have worked on a Roman rural estate.
The researchers mined tiny DNA fragments from dental pulp. They were able to extract, purify, and enrich specifically for the Plasmodium species known to infect humans.
The team noted that usable DNA is challenging to extract because the parasites primarily dwell within the bloodstream and organs, which decompose and break down over time—in this instance, over the course of 2 millennia.
However, the researchers recovered more than half of the P falciparum mitochondrial genome from 2 individuals from Velia and Vagnari. ![]()

individual from Velia, Italy
Photo courtesy of Luca
Bandioli, Pigorini Museum
An analysis of 2000-year-old human remains from several regions across the Italian peninsula has confirmed the presence of Plasmodium falciparum malaria during the Roman Empire, according to researchers.
The team found mitochondrial genomic evidence of P falciparum malaria, coaxed from the teeth of bodies buried in 3 Italian cemeteries, dating back to the Imperial period.
The researchers said these finding provide a key reference point for when and where the malaria parasite existed in humans, as well as more information about the evolution of human disease.
The team reported these findings in Current Biology.
“There is extensive written evidence describing fevers that sound like malaria in ancient Greece and Rome, but the specific malaria species responsible is unknown,” said study author Stephanie Marciniak, PhD, of Pennsylvania State University in University Park.
“Our data confirm that the species was likely Plasmodium falciparum and that it affected people in different ecological and cultural environments. These results open up new questions to explore, particularly how widespread this parasite was and what burden it placed upon communities in Imperial Roman Italy.”
Dr Marciniak and her colleagues sampled teeth taken from 58 adults interred at 3 Imperial period Italian cemeteries: Isola Sacra, Velia, and Vagnari.
Located on the coast, Velia and Isola Sacra were known as important port cities and trading centers. Vagnari is located further inland and believed to be the burial site of laborers who would have worked on a Roman rural estate.
The researchers mined tiny DNA fragments from dental pulp. They were able to extract, purify, and enrich specifically for the Plasmodium species known to infect humans.
The team noted that usable DNA is challenging to extract because the parasites primarily dwell within the bloodstream and organs, which decompose and break down over time—in this instance, over the course of 2 millennia.
However, the researchers recovered more than half of the P falciparum mitochondrial genome from 2 individuals from Velia and Vagnari. ![]()
Group estimates global cancer cases, deaths in 2015
receiving chemotherapy
Photo by Rhoda Baer
Researchers have estimated the global incidence of 32 cancer types and deaths related to these malignancies in 2015.
The group’s data, published in JAMA Oncology, suggest there were 17.5 million cancer cases and 8.7 million cancer deaths last year.
There were 78,000 cases of Hodgkin lymphoma and 24,000 deaths from the disease, as well as 666,000 cases of non-Hodgkin lymphoma (NHL) and 231,000 NHL deaths.
There were 154,000 cases of multiple myeloma and 101,000 deaths from the disease.
And there were 606,000 cases of leukemia, with 353,000 leukemia deaths. This included 161,000 cases of acute lymphoid leukemia (110,000 deaths), 191,000 cases of chronic lymphoid leukemia (61,000 deaths), 190,000 cases of acute myeloid leukemia (147,000 deaths), and 64,000 cases of chronic myeloid leukemia (35,000 deaths).
The data also show that, between 2005 and 2015, cancer cases increased by 33%, mostly due to population aging and growth, plus changes in age-specific cancer rates.
Globally, the odds of developing cancer during a lifetime were 1 in 3 for men and 1 in 4 for women in 2015.
Prostate cancer was the most common cancer in men (1.6 million cases), although tracheal, bronchus, and lung cancer was the leading cause of cancer deaths for men.
Breast cancer was the most common cancer for women (2.4 million cases) and the leading cause of cancer deaths in women.
The most common childhood cancers were leukemia, “other neoplasms,” NHL, and brain and nervous system cancers. ![]()

receiving chemotherapy
Photo by Rhoda Baer
Researchers have estimated the global incidence of 32 cancer types and deaths related to these malignancies in 2015.
The group’s data, published in JAMA Oncology, suggest there were 17.5 million cancer cases and 8.7 million cancer deaths last year.
There were 78,000 cases of Hodgkin lymphoma and 24,000 deaths from the disease, as well as 666,000 cases of non-Hodgkin lymphoma (NHL) and 231,000 NHL deaths.
There were 154,000 cases of multiple myeloma and 101,000 deaths from the disease.
And there were 606,000 cases of leukemia, with 353,000 leukemia deaths. This included 161,000 cases of acute lymphoid leukemia (110,000 deaths), 191,000 cases of chronic lymphoid leukemia (61,000 deaths), 190,000 cases of acute myeloid leukemia (147,000 deaths), and 64,000 cases of chronic myeloid leukemia (35,000 deaths).
The data also show that, between 2005 and 2015, cancer cases increased by 33%, mostly due to population aging and growth, plus changes in age-specific cancer rates.
Globally, the odds of developing cancer during a lifetime were 1 in 3 for men and 1 in 4 for women in 2015.
Prostate cancer was the most common cancer in men (1.6 million cases), although tracheal, bronchus, and lung cancer was the leading cause of cancer deaths for men.
Breast cancer was the most common cancer for women (2.4 million cases) and the leading cause of cancer deaths in women.
The most common childhood cancers were leukemia, “other neoplasms,” NHL, and brain and nervous system cancers. ![]()

receiving chemotherapy
Photo by Rhoda Baer
Researchers have estimated the global incidence of 32 cancer types and deaths related to these malignancies in 2015.
The group’s data, published in JAMA Oncology, suggest there were 17.5 million cancer cases and 8.7 million cancer deaths last year.
There were 78,000 cases of Hodgkin lymphoma and 24,000 deaths from the disease, as well as 666,000 cases of non-Hodgkin lymphoma (NHL) and 231,000 NHL deaths.
There were 154,000 cases of multiple myeloma and 101,000 deaths from the disease.
And there were 606,000 cases of leukemia, with 353,000 leukemia deaths. This included 161,000 cases of acute lymphoid leukemia (110,000 deaths), 191,000 cases of chronic lymphoid leukemia (61,000 deaths), 190,000 cases of acute myeloid leukemia (147,000 deaths), and 64,000 cases of chronic myeloid leukemia (35,000 deaths).
The data also show that, between 2005 and 2015, cancer cases increased by 33%, mostly due to population aging and growth, plus changes in age-specific cancer rates.
Globally, the odds of developing cancer during a lifetime were 1 in 3 for men and 1 in 4 for women in 2015.
Prostate cancer was the most common cancer in men (1.6 million cases), although tracheal, bronchus, and lung cancer was the leading cause of cancer deaths for men.
Breast cancer was the most common cancer for women (2.4 million cases) and the leading cause of cancer deaths in women.
The most common childhood cancers were leukemia, “other neoplasms,” NHL, and brain and nervous system cancers. ![]()
Malaria elimination in sub-Saharan Africa is possible, study suggests

Kariba area of southern Zambia
where malaria elimination
programs are underway.
Photo from Milen Nikolov
Malaria elimination in historically high transmission areas like southern Africa is possible with tools that are already available, according to new research.
The study suggests that high levels of vector control are key, and mass drug campaigns cannot make much of an impact without proper vector control.
Milen Nikolov, of the Institute for Disease Modeling in Bellevue, Washington, and colleagues reported these findings in PLOS Computational Biology.
The researchers said the Lake Kariba region of Southern Province, Zambia, is part of a multi-country malaria elimination effort. However, elimination in this area is challenging because villages with high and low malaria burden are interconnected through human travel.
With this in mind, the researchers combined a mathematical model of malaria transmission with field data from Zambia to test a variety of strategies for eliminating malaria in the Lake Kariba region.
The team used detailed spatial surveillance data from field studies—including household locations, climate, clinical malaria incidence, prevalence of malaria infections, and bednet usage rates—to construct a model of interconnected villages, then tested a variety of intervention scenarios to see which ones could lead to elimination.
The results indicate that elimination requires high, yet realistic, levels of vector control. And mass drug campaigns deployed to kill parasites in the human population can boost the chances of achieving elimination as long as vector control is well-implemented.
The researchers said this work suggests that elimination programs in sub-Saharan Africa should focus on how to achieve and maintain excellent coverage of vector control measures rather than spending resources on mass drug campaigns that are predicted to have little effect without well-implemented vector control already in place.
Human movement within the region should be targeted to achieve elimination, as should the importation of infections from outside the region. This is because both impact the likelihood of achieving elimination and understanding regional movement patterns can help guide strategies on targeting specific groups of at-risk people.
While no sub-Saharan African country has yet eliminated malaria, the researchers predict that regional malaria elimination is within reach with current tools, provided the efficacy and operational efficiency attained in southern Zambia can be extended and targeted to other key areas. ![]()

Kariba area of southern Zambia
where malaria elimination
programs are underway.
Photo from Milen Nikolov
Malaria elimination in historically high transmission areas like southern Africa is possible with tools that are already available, according to new research.
The study suggests that high levels of vector control are key, and mass drug campaigns cannot make much of an impact without proper vector control.
Milen Nikolov, of the Institute for Disease Modeling in Bellevue, Washington, and colleagues reported these findings in PLOS Computational Biology.
The researchers said the Lake Kariba region of Southern Province, Zambia, is part of a multi-country malaria elimination effort. However, elimination in this area is challenging because villages with high and low malaria burden are interconnected through human travel.
With this in mind, the researchers combined a mathematical model of malaria transmission with field data from Zambia to test a variety of strategies for eliminating malaria in the Lake Kariba region.
The team used detailed spatial surveillance data from field studies—including household locations, climate, clinical malaria incidence, prevalence of malaria infections, and bednet usage rates—to construct a model of interconnected villages, then tested a variety of intervention scenarios to see which ones could lead to elimination.
The results indicate that elimination requires high, yet realistic, levels of vector control. And mass drug campaigns deployed to kill parasites in the human population can boost the chances of achieving elimination as long as vector control is well-implemented.
The researchers said this work suggests that elimination programs in sub-Saharan Africa should focus on how to achieve and maintain excellent coverage of vector control measures rather than spending resources on mass drug campaigns that are predicted to have little effect without well-implemented vector control already in place.
Human movement within the region should be targeted to achieve elimination, as should the importation of infections from outside the region. This is because both impact the likelihood of achieving elimination and understanding regional movement patterns can help guide strategies on targeting specific groups of at-risk people.
While no sub-Saharan African country has yet eliminated malaria, the researchers predict that regional malaria elimination is within reach with current tools, provided the efficacy and operational efficiency attained in southern Zambia can be extended and targeted to other key areas. ![]()

Kariba area of southern Zambia
where malaria elimination
programs are underway.
Photo from Milen Nikolov
Malaria elimination in historically high transmission areas like southern Africa is possible with tools that are already available, according to new research.
The study suggests that high levels of vector control are key, and mass drug campaigns cannot make much of an impact without proper vector control.
Milen Nikolov, of the Institute for Disease Modeling in Bellevue, Washington, and colleagues reported these findings in PLOS Computational Biology.
The researchers said the Lake Kariba region of Southern Province, Zambia, is part of a multi-country malaria elimination effort. However, elimination in this area is challenging because villages with high and low malaria burden are interconnected through human travel.
With this in mind, the researchers combined a mathematical model of malaria transmission with field data from Zambia to test a variety of strategies for eliminating malaria in the Lake Kariba region.
The team used detailed spatial surveillance data from field studies—including household locations, climate, clinical malaria incidence, prevalence of malaria infections, and bednet usage rates—to construct a model of interconnected villages, then tested a variety of intervention scenarios to see which ones could lead to elimination.
The results indicate that elimination requires high, yet realistic, levels of vector control. And mass drug campaigns deployed to kill parasites in the human population can boost the chances of achieving elimination as long as vector control is well-implemented.
The researchers said this work suggests that elimination programs in sub-Saharan Africa should focus on how to achieve and maintain excellent coverage of vector control measures rather than spending resources on mass drug campaigns that are predicted to have little effect without well-implemented vector control already in place.
Human movement within the region should be targeted to achieve elimination, as should the importation of infections from outside the region. This is because both impact the likelihood of achieving elimination and understanding regional movement patterns can help guide strategies on targeting specific groups of at-risk people.
While no sub-Saharan African country has yet eliminated malaria, the researchers predict that regional malaria elimination is within reach with current tools, provided the efficacy and operational efficiency attained in southern Zambia can be extended and targeted to other key areas. ![]()
FDA authorizes emergency use of Zika assay

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has issued an emergency use authorization (EUA) for Abbott Molecular Inc.’s RealTime ZIKA assay.
The EUA means the assay can be used by certified laboratories for the qualitative detection of RNA from Zika virus in human serum, EDTA plasma, and urine (collected alongside a patient-matched serum or plasma specimen).
Zika virus RNA is generally detectable in these specimens during the acute phase of infection.
According to updated guidance from the US Centers for Disease Control and Prevention (CDC), Zika virus RNA is detectable up to 14 days in serum and urine (possibly longer in urine), following the onset of symptoms, if present. Positive results are indicative of current infection.
The FDA’s decision to grant an EUA means Abbott’s RealTime ZIKA assay can be used in individuals who meet CDC Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiological criteria for which Zika virus testing may be indicated).
The assay can be used by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high complexity tests, or by similarly qualified non-US laboratories, pursuant to section 564 of the Federal Food, Drug, and Cosmetic Act (21 U.S.C. § 360bbb-3).
The EUA does not mean Abbott’s RealTime ZIKA assay is FDA cleared or approved.
An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means Abbott’s RealTime ZIKA assay is only authorized as long as circumstances exist to justify the emergency use of in vitro diagnostics for the detection of Zika virus, unless the authorization is terminated or revoked sooner. ![]()

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has issued an emergency use authorization (EUA) for Abbott Molecular Inc.’s RealTime ZIKA assay.
The EUA means the assay can be used by certified laboratories for the qualitative detection of RNA from Zika virus in human serum, EDTA plasma, and urine (collected alongside a patient-matched serum or plasma specimen).
Zika virus RNA is generally detectable in these specimens during the acute phase of infection.
According to updated guidance from the US Centers for Disease Control and Prevention (CDC), Zika virus RNA is detectable up to 14 days in serum and urine (possibly longer in urine), following the onset of symptoms, if present. Positive results are indicative of current infection.
The FDA’s decision to grant an EUA means Abbott’s RealTime ZIKA assay can be used in individuals who meet CDC Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiological criteria for which Zika virus testing may be indicated).
The assay can be used by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high complexity tests, or by similarly qualified non-US laboratories, pursuant to section 564 of the Federal Food, Drug, and Cosmetic Act (21 U.S.C. § 360bbb-3).
The EUA does not mean Abbott’s RealTime ZIKA assay is FDA cleared or approved.
An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means Abbott’s RealTime ZIKA assay is only authorized as long as circumstances exist to justify the emergency use of in vitro diagnostics for the detection of Zika virus, unless the authorization is terminated or revoked sooner. ![]()

Photo by Juan D. Alfonso
The US Food and Drug Administration (FDA) has issued an emergency use authorization (EUA) for Abbott Molecular Inc.’s RealTime ZIKA assay.
The EUA means the assay can be used by certified laboratories for the qualitative detection of RNA from Zika virus in human serum, EDTA plasma, and urine (collected alongside a patient-matched serum or plasma specimen).
Zika virus RNA is generally detectable in these specimens during the acute phase of infection.
According to updated guidance from the US Centers for Disease Control and Prevention (CDC), Zika virus RNA is detectable up to 14 days in serum and urine (possibly longer in urine), following the onset of symptoms, if present. Positive results are indicative of current infection.
The FDA’s decision to grant an EUA means Abbott’s RealTime ZIKA assay can be used in individuals who meet CDC Zika virus clinical criteria (eg, clinical signs and symptoms associated with Zika virus infection) and/or CDC Zika virus epidemiological criteria (eg, history of residence in or travel to a geographic region with active Zika transmission at the time of travel, or other epidemiological criteria for which Zika virus testing may be indicated).
The assay can be used by laboratories in the US that are certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high complexity tests, or by similarly qualified non-US laboratories, pursuant to section 564 of the Federal Food, Drug, and Cosmetic Act (21 U.S.C. § 360bbb-3).
The EUA does not mean Abbott’s RealTime ZIKA assay is FDA cleared or approved.
An EUA allows for the use of unapproved medical products or unapproved uses of approved medical products in an emergency.
The products must be used to diagnose, treat, or prevent serious or life-threatening conditions caused by chemical, biological, radiological, or nuclear threat agents, when there are no adequate alternatives.
This means Abbott’s RealTime ZIKA assay is only authorized as long as circumstances exist to justify the emergency use of in vitro diagnostics for the detection of Zika virus, unless the authorization is terminated or revoked sooner. ![]()
NCCN releases new guidelines for cancer patients

Nausea and Vomiting
©NCCN® 2016
The National Comprehensive Cancer Network (NCCN) has released new educational materials designed to help cancer patients combat nausea and vomiting.
The NCCN Guidelines for Patients® for Nausea and Vomiting and NCCN Quick Guide™ for Nausea and Vomiting are the first patient resources from NCCN to focus specifically on supportive care.
The resources are available on NCCN.org/patients and via the NCCN Patient Guides for Cancer mobile app.
NCCN Guidelines for Patients are patient-friendly translations of the NCCN Clinical Practice Guidelines in Oncology. Each resource features guidance from US cancer centers designed to help people living with cancer talk with their physicians about the best treatment options for their disease.
NCCN Quick Guide™ sheets are 1-page summaries of key points in the patient guidelines. They include elements such as “questions to ask your doctor,” a glossary of terms, and medical illustrations of anatomy, tests, and treatments.
The NCCN Guidelines for Patients for Nausea and Vomiting:
- Explain how these side effects are related to cancer treatment
- List cancer treatments that can cause nausea and vomiting
- Detail methods of preventing and treating these side effects
- Outline methods of coping with nausea and vomiting
- Provide a list of resources for information and support.
“At NCCN, our mission is to improve the lives of patients with cancer, and we are excited to be able to provide the information that will help patients better understand this common side effect of cancer treatment,” said Marcie R. Reeder, executive director of the NCCN Foundation.
“The NCCN Guidelines for Patients for Nausea and Vomiting are the first of a highly anticipated library of supportive care resources that provide patients with the same information their doctors use.” ![]()

Nausea and Vomiting
©NCCN® 2016
The National Comprehensive Cancer Network (NCCN) has released new educational materials designed to help cancer patients combat nausea and vomiting.
The NCCN Guidelines for Patients® for Nausea and Vomiting and NCCN Quick Guide™ for Nausea and Vomiting are the first patient resources from NCCN to focus specifically on supportive care.
The resources are available on NCCN.org/patients and via the NCCN Patient Guides for Cancer mobile app.
NCCN Guidelines for Patients are patient-friendly translations of the NCCN Clinical Practice Guidelines in Oncology. Each resource features guidance from US cancer centers designed to help people living with cancer talk with their physicians about the best treatment options for their disease.
NCCN Quick Guide™ sheets are 1-page summaries of key points in the patient guidelines. They include elements such as “questions to ask your doctor,” a glossary of terms, and medical illustrations of anatomy, tests, and treatments.
The NCCN Guidelines for Patients for Nausea and Vomiting:
- Explain how these side effects are related to cancer treatment
- List cancer treatments that can cause nausea and vomiting
- Detail methods of preventing and treating these side effects
- Outline methods of coping with nausea and vomiting
- Provide a list of resources for information and support.
“At NCCN, our mission is to improve the lives of patients with cancer, and we are excited to be able to provide the information that will help patients better understand this common side effect of cancer treatment,” said Marcie R. Reeder, executive director of the NCCN Foundation.
“The NCCN Guidelines for Patients for Nausea and Vomiting are the first of a highly anticipated library of supportive care resources that provide patients with the same information their doctors use.” ![]()

Nausea and Vomiting
©NCCN® 2016
The National Comprehensive Cancer Network (NCCN) has released new educational materials designed to help cancer patients combat nausea and vomiting.
The NCCN Guidelines for Patients® for Nausea and Vomiting and NCCN Quick Guide™ for Nausea and Vomiting are the first patient resources from NCCN to focus specifically on supportive care.
The resources are available on NCCN.org/patients and via the NCCN Patient Guides for Cancer mobile app.
NCCN Guidelines for Patients are patient-friendly translations of the NCCN Clinical Practice Guidelines in Oncology. Each resource features guidance from US cancer centers designed to help people living with cancer talk with their physicians about the best treatment options for their disease.
NCCN Quick Guide™ sheets are 1-page summaries of key points in the patient guidelines. They include elements such as “questions to ask your doctor,” a glossary of terms, and medical illustrations of anatomy, tests, and treatments.
The NCCN Guidelines for Patients for Nausea and Vomiting:
- Explain how these side effects are related to cancer treatment
- List cancer treatments that can cause nausea and vomiting
- Detail methods of preventing and treating these side effects
- Outline methods of coping with nausea and vomiting
- Provide a list of resources for information and support.
“At NCCN, our mission is to improve the lives of patients with cancer, and we are excited to be able to provide the information that will help patients better understand this common side effect of cancer treatment,” said Marcie R. Reeder, executive director of the NCCN Foundation.
“The NCCN Guidelines for Patients for Nausea and Vomiting are the first of a highly anticipated library of supportive care resources that provide patients with the same information their doctors use.” ![]()
Large-scale tumor profiling deemed feasible, but challenges remain

Photo courtesy of the
National Institute of
General Medical Sciences
New research suggests large-scale genomic profiling is technically feasible in a broad population of cancer patients.
However, the study also revealed challenges and barriers to widespread implementation of precision medicine, according to researchers.
Specifically, half of the patients studied did not receive results of genomic profiling due to insufficient samples or sequencing failure.
Most patients who did receive results did not see a change in their care.
However, genomic profiling provided an accurate diagnosis and changed treatment for a handful of the patients studied.
Lynette M. Sholl, MD, of Brigham and Women’s Hospital in Boston, Massachusetts, and her colleagues reported these findings in JCI Insight.
The report contains data on pediatric and adult patients with a range of malignancies.
Patient samples were analyzed using OncoPanel. This platform sequences hundreds of known cancer-related genes to look for alterations that drive tumors and might be helpful in guiding treatment choice or enrolling the patient in an appropriate clinical trial.
This study began with 7397 patients, but many of these individuals did not have specimens adequate for sequencing. This left 3892 patients (53%) to undergo genomic profiling, but sequencing failed in 165 (4%) of them. So sequencing was successful in 3727 patients, or 50% of the overall population.
Of the 3727 patients in whom sequencing was successful, 73% had at least 1 genetic alteration that was considered “clinically actionable or informative” by the researchers.
This included 54% of patients with alterations that might be used to inform diagnosis or recommend enrollment in a clinical trial. It also included 19% of patients who had an alteration that “would inform standard-of-care therapeutic decision-making,” according to the researchers.
The team provided several examples of how genomic testing clarified or changed a patient’s diagnosis, which, in turn, altered treatment and prognosis.
One example was a patient who was originally diagnosed with peripheral T-cell lymphoma, which was later revised to myeloid sarcoma. Sequencing results suggested the patient actually had FIP1L1-PDGFRA-driven acute myeloid leukemia, which predicted responsiveness to imatinib.
The patient was treated with imatinib and experienced a “dramatic and sustained clinical response.” He then proceeded to allogeneic transplant and had no evidence of disease at 1 year of follow-up.
The researchers concluded that genomic sequencing results may alter the management of cancer patients in some cases, but certain barriers must be overcome to enable precision cancer medicine on a large scale. ![]()

Photo courtesy of the
National Institute of
General Medical Sciences
New research suggests large-scale genomic profiling is technically feasible in a broad population of cancer patients.
However, the study also revealed challenges and barriers to widespread implementation of precision medicine, according to researchers.
Specifically, half of the patients studied did not receive results of genomic profiling due to insufficient samples or sequencing failure.
Most patients who did receive results did not see a change in their care.
However, genomic profiling provided an accurate diagnosis and changed treatment for a handful of the patients studied.
Lynette M. Sholl, MD, of Brigham and Women’s Hospital in Boston, Massachusetts, and her colleagues reported these findings in JCI Insight.
The report contains data on pediatric and adult patients with a range of malignancies.
Patient samples were analyzed using OncoPanel. This platform sequences hundreds of known cancer-related genes to look for alterations that drive tumors and might be helpful in guiding treatment choice or enrolling the patient in an appropriate clinical trial.
This study began with 7397 patients, but many of these individuals did not have specimens adequate for sequencing. This left 3892 patients (53%) to undergo genomic profiling, but sequencing failed in 165 (4%) of them. So sequencing was successful in 3727 patients, or 50% of the overall population.
Of the 3727 patients in whom sequencing was successful, 73% had at least 1 genetic alteration that was considered “clinically actionable or informative” by the researchers.
This included 54% of patients with alterations that might be used to inform diagnosis or recommend enrollment in a clinical trial. It also included 19% of patients who had an alteration that “would inform standard-of-care therapeutic decision-making,” according to the researchers.
The team provided several examples of how genomic testing clarified or changed a patient’s diagnosis, which, in turn, altered treatment and prognosis.
One example was a patient who was originally diagnosed with peripheral T-cell lymphoma, which was later revised to myeloid sarcoma. Sequencing results suggested the patient actually had FIP1L1-PDGFRA-driven acute myeloid leukemia, which predicted responsiveness to imatinib.
The patient was treated with imatinib and experienced a “dramatic and sustained clinical response.” He then proceeded to allogeneic transplant and had no evidence of disease at 1 year of follow-up.
The researchers concluded that genomic sequencing results may alter the management of cancer patients in some cases, but certain barriers must be overcome to enable precision cancer medicine on a large scale. ![]()

Photo courtesy of the
National Institute of
General Medical Sciences
New research suggests large-scale genomic profiling is technically feasible in a broad population of cancer patients.
However, the study also revealed challenges and barriers to widespread implementation of precision medicine, according to researchers.
Specifically, half of the patients studied did not receive results of genomic profiling due to insufficient samples or sequencing failure.
Most patients who did receive results did not see a change in their care.
However, genomic profiling provided an accurate diagnosis and changed treatment for a handful of the patients studied.
Lynette M. Sholl, MD, of Brigham and Women’s Hospital in Boston, Massachusetts, and her colleagues reported these findings in JCI Insight.
The report contains data on pediatric and adult patients with a range of malignancies.
Patient samples were analyzed using OncoPanel. This platform sequences hundreds of known cancer-related genes to look for alterations that drive tumors and might be helpful in guiding treatment choice or enrolling the patient in an appropriate clinical trial.
This study began with 7397 patients, but many of these individuals did not have specimens adequate for sequencing. This left 3892 patients (53%) to undergo genomic profiling, but sequencing failed in 165 (4%) of them. So sequencing was successful in 3727 patients, or 50% of the overall population.
Of the 3727 patients in whom sequencing was successful, 73% had at least 1 genetic alteration that was considered “clinically actionable or informative” by the researchers.
This included 54% of patients with alterations that might be used to inform diagnosis or recommend enrollment in a clinical trial. It also included 19% of patients who had an alteration that “would inform standard-of-care therapeutic decision-making,” according to the researchers.
The team provided several examples of how genomic testing clarified or changed a patient’s diagnosis, which, in turn, altered treatment and prognosis.
One example was a patient who was originally diagnosed with peripheral T-cell lymphoma, which was later revised to myeloid sarcoma. Sequencing results suggested the patient actually had FIP1L1-PDGFRA-driven acute myeloid leukemia, which predicted responsiveness to imatinib.
The patient was treated with imatinib and experienced a “dramatic and sustained clinical response.” He then proceeded to allogeneic transplant and had no evidence of disease at 1 year of follow-up.
The researchers concluded that genomic sequencing results may alter the management of cancer patients in some cases, but certain barriers must be overcome to enable precision cancer medicine on a large scale.
Device provides long-lasting drug delivery

with 6 arms that can be folded
and encased in a capsule.
Photo by Melanie Gonick/MIT
A new device can provide long-term, controlled drug release, according to research published in Science Translational Medicine.
Researchers tested this device—a 6-armed structure encased in a capsule—by loading it with ivermectin, an antiparasitic drug that disrupts malaria transmission by killing infected mosquitoes.
When administered to pigs, the device safely stayed in the stomach, slowly releasing ivermectin for up to 14 days.
Researchers believe this type of lasting drug delivery could help bolster the success of malaria elimination campaigns, which rely on treatment adherence and cost-effective approaches that reach large, rural populations.
The team also believes this device could be used to treat a range of other diseases, particularly those in which treatment adherence may be an issue.
“We want to make it as easy as possible for people to take their medications over a sustained period of time,” said study author C. Giovanni Traverso, MB, BChir, PhD, of the Massachusetts Institute of Technology (MIT) in Cambridge.
“When patients have to remember to take a drug every day or multiple times a day, we start to see less and less adherence to the regimen. Being able to swallow a capsule once a week or once a month could change the way we think about delivering medications.”
This research has led to the launch of Lyndra, a company that is developing this technology with a focus on diseases for which patients would benefit the most from sustained drug delivery. This includes neuropsychiatric disorders, HIV, diabetes, and epilepsy.
Designing, testing the device
To provide long-term drug delivery, the researchers designed a star-shaped device with 6 arms that can be folded inward and encased in a smooth capsule.
Drug molecules are loaded into the arms, which are made of a rigid polymer called polycaprolactone. Each arm is attached to a rubber-like core by a linker that is designed to eventually break down.
After the capsule is swallowed, stomach acid dissolves the outer layer, allowing the 6-armed device to unfold.
Once the device expands, it is large enough to stay in the stomach and resist the forces that would normally push an object further down the digestive tract. However, it is not large enough to cause any harmful blockage of the digestive tract.
The drug is gradually released over a period of several days. After that, the linkers that join the arms to the core of the device dissolve, allowing the arms to break off. The pieces are small enough that they can pass harmlessly through the digestive tract.
In pigs, the device slowly released ivermectin for up to 14 days without causing injury to the stomach or obstructing the passage of food, before breaking apart and passing safely out of the body.
Modeling the impact
The researchers used mathematical modeling to predict the potential impact of this drug delivery method.
The data suggested that if the device were used to deliver ivermectin, it could increase the efficacy of mass drug administration campaigns designed to fight malaria.
“What we showed is that we stand to significantly amplify the effect of those campaigns,” Dr Traverso said. “The introduction of this kind of system could have a substantial impact on the fight against malaria and transform clinical care in general by ensuring patients receive their medication.”
Potential applications
“Until now, oral drugs would almost never last for more than a day,” said study author Robert Langer, ScD, of MIT.
“This really opens the door to ultra-long-lasting oral systems, which could have an effect on all kinds of diseases, such as Alzheimer’s or mental health disorders. There are a lot of exciting things this could someday enable.”
“This is a platform into which you can incorporate any drug,” added Mousa Jafari, PhD, of MIT. “This can be used with any drug that requires frequent dosing. We can replace that dosing with a single administration.”
This type of delivery could also help researchers run better clinical trials by making it easier for patients to take the drugs, said Shiyi Zhang, PhD, of MIT.
“It may help doctors and the pharma industry to better evaluate the efficacy of certain drugs because, currently, a lot of patients in clinical trials have serious medication adherence problems that will mislead the clinical studies,” he said.

with 6 arms that can be folded
and encased in a capsule.
Photo by Melanie Gonick/MIT
A new device can provide long-term, controlled drug release, according to research published in Science Translational Medicine.
Researchers tested this device—a 6-armed structure encased in a capsule—by loading it with ivermectin, an antiparasitic drug that disrupts malaria transmission by killing infected mosquitoes.
When administered to pigs, the device safely stayed in the stomach, slowly releasing ivermectin for up to 14 days.
Researchers believe this type of lasting drug delivery could help bolster the success of malaria elimination campaigns, which rely on treatment adherence and cost-effective approaches that reach large, rural populations.
The team also believes this device could be used to treat a range of other diseases, particularly those in which treatment adherence may be an issue.
“We want to make it as easy as possible for people to take their medications over a sustained period of time,” said study author C. Giovanni Traverso, MB, BChir, PhD, of the Massachusetts Institute of Technology (MIT) in Cambridge.
“When patients have to remember to take a drug every day or multiple times a day, we start to see less and less adherence to the regimen. Being able to swallow a capsule once a week or once a month could change the way we think about delivering medications.”
This research has led to the launch of Lyndra, a company that is developing this technology with a focus on diseases for which patients would benefit the most from sustained drug delivery. This includes neuropsychiatric disorders, HIV, diabetes, and epilepsy.
Designing, testing the device
To provide long-term drug delivery, the researchers designed a star-shaped device with 6 arms that can be folded inward and encased in a smooth capsule.
Drug molecules are loaded into the arms, which are made of a rigid polymer called polycaprolactone. Each arm is attached to a rubber-like core by a linker that is designed to eventually break down.
After the capsule is swallowed, stomach acid dissolves the outer layer, allowing the 6-armed device to unfold.
Once the device expands, it is large enough to stay in the stomach and resist the forces that would normally push an object further down the digestive tract. However, it is not large enough to cause any harmful blockage of the digestive tract.
The drug is gradually released over a period of several days. After that, the linkers that join the arms to the core of the device dissolve, allowing the arms to break off. The pieces are small enough that they can pass harmlessly through the digestive tract.
In pigs, the device slowly released ivermectin for up to 14 days without causing injury to the stomach or obstructing the passage of food, before breaking apart and passing safely out of the body.
Modeling the impact
The researchers used mathematical modeling to predict the potential impact of this drug delivery method.
The data suggested that if the device were used to deliver ivermectin, it could increase the efficacy of mass drug administration campaigns designed to fight malaria.
“What we showed is that we stand to significantly amplify the effect of those campaigns,” Dr Traverso said. “The introduction of this kind of system could have a substantial impact on the fight against malaria and transform clinical care in general by ensuring patients receive their medication.”
Potential applications
“Until now, oral drugs would almost never last for more than a day,” said study author Robert Langer, ScD, of MIT.
“This really opens the door to ultra-long-lasting oral systems, which could have an effect on all kinds of diseases, such as Alzheimer’s or mental health disorders. There are a lot of exciting things this could someday enable.”
“This is a platform into which you can incorporate any drug,” added Mousa Jafari, PhD, of MIT. “This can be used with any drug that requires frequent dosing. We can replace that dosing with a single administration.”
This type of delivery could also help researchers run better clinical trials by making it easier for patients to take the drugs, said Shiyi Zhang, PhD, of MIT.
“It may help doctors and the pharma industry to better evaluate the efficacy of certain drugs because, currently, a lot of patients in clinical trials have serious medication adherence problems that will mislead the clinical studies,” he said.

with 6 arms that can be folded
and encased in a capsule.
Photo by Melanie Gonick/MIT
A new device can provide long-term, controlled drug release, according to research published in Science Translational Medicine.
Researchers tested this device—a 6-armed structure encased in a capsule—by loading it with ivermectin, an antiparasitic drug that disrupts malaria transmission by killing infected mosquitoes.
When administered to pigs, the device safely stayed in the stomach, slowly releasing ivermectin for up to 14 days.
Researchers believe this type of lasting drug delivery could help bolster the success of malaria elimination campaigns, which rely on treatment adherence and cost-effective approaches that reach large, rural populations.
The team also believes this device could be used to treat a range of other diseases, particularly those in which treatment adherence may be an issue.
“We want to make it as easy as possible for people to take their medications over a sustained period of time,” said study author C. Giovanni Traverso, MB, BChir, PhD, of the Massachusetts Institute of Technology (MIT) in Cambridge.
“When patients have to remember to take a drug every day or multiple times a day, we start to see less and less adherence to the regimen. Being able to swallow a capsule once a week or once a month could change the way we think about delivering medications.”
This research has led to the launch of Lyndra, a company that is developing this technology with a focus on diseases for which patients would benefit the most from sustained drug delivery. This includes neuropsychiatric disorders, HIV, diabetes, and epilepsy.
Designing, testing the device
To provide long-term drug delivery, the researchers designed a star-shaped device with 6 arms that can be folded inward and encased in a smooth capsule.
Drug molecules are loaded into the arms, which are made of a rigid polymer called polycaprolactone. Each arm is attached to a rubber-like core by a linker that is designed to eventually break down.
After the capsule is swallowed, stomach acid dissolves the outer layer, allowing the 6-armed device to unfold.
Once the device expands, it is large enough to stay in the stomach and resist the forces that would normally push an object further down the digestive tract. However, it is not large enough to cause any harmful blockage of the digestive tract.
The drug is gradually released over a period of several days. After that, the linkers that join the arms to the core of the device dissolve, allowing the arms to break off. The pieces are small enough that they can pass harmlessly through the digestive tract.
In pigs, the device slowly released ivermectin for up to 14 days without causing injury to the stomach or obstructing the passage of food, before breaking apart and passing safely out of the body.
Modeling the impact
The researchers used mathematical modeling to predict the potential impact of this drug delivery method.
The data suggested that if the device were used to deliver ivermectin, it could increase the efficacy of mass drug administration campaigns designed to fight malaria.
“What we showed is that we stand to significantly amplify the effect of those campaigns,” Dr Traverso said. “The introduction of this kind of system could have a substantial impact on the fight against malaria and transform clinical care in general by ensuring patients receive their medication.”
Potential applications
“Until now, oral drugs would almost never last for more than a day,” said study author Robert Langer, ScD, of MIT.
“This really opens the door to ultra-long-lasting oral systems, which could have an effect on all kinds of diseases, such as Alzheimer’s or mental health disorders. There are a lot of exciting things this could someday enable.”
“This is a platform into which you can incorporate any drug,” added Mousa Jafari, PhD, of MIT. “This can be used with any drug that requires frequent dosing. We can replace that dosing with a single administration.”
This type of delivery could also help researchers run better clinical trials by making it easier for patients to take the drugs, said Shiyi Zhang, PhD, of MIT.
“It may help doctors and the pharma industry to better evaluate the efficacy of certain drugs because, currently, a lot of patients in clinical trials have serious medication adherence problems that will mislead the clinical studies,” he said.
Mutations aid resistance, growth of malaria parasite
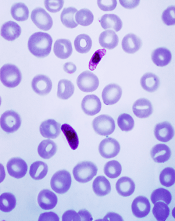
Plasmodium falciparum
Image from CDC/Mae Melvin
Some mutations that enable drug resistance in the malaria parasite Plasmodium falciparum may also help it grow, according to a study published in PLOS Pathogens.
Some strains of P falciparum have evolved to become resistant to antimalarial drugs, including chloroquine.
Often, chloroquine resistance mutations hinder P falciparum’s ability to infect the bloodstream and grow.
However, in a previous study, researchers discovered that a uniquely mutated version of the P falciparum gene pfcrt provides drug resistance while avoiding the detrimental impact of growth seen with other mutated pfcrt variants.
In the new study, the same group of researchers—Stanislaw Gabryszewski, of Columbia University Medical Center in New York, and his colleagues—investigated this version of the pfcrt gene, which is called Cam734 and has been found in certain regions in Southeast Asia.
Using zinc-finger nucleases, the team characterized the individual mutations unique to Cam734 in terms of their effects on drug resistance, metabolism, and growth rates in living parasites.
The researchers found that a mutation called A144F is required for the chloroquine resistance enabled by Cam734, and this mutation also contributes to resistance to the drugs amodiaquine and quinine.
The team identified additional mutations that contribute to resistance to chloroquine and impact the potency of other antimalarials as well.
When the researchers reversed these mutations in living parasites that had the Cam734 allele, growth slowed, indicating that these mutations also enhance infection.
Additional experiments revealed specific effects of Cam734 mutations on several metabolic pathways in P falciparum, including the digestion of human hemoglobin that parasites use to obtain amino acids for protein synthesis.
The researchers also found evidence suggesting that Cam734 helps to maintain an electrochemical gradient that allows the protein encoded by the pfcrt gene to thwart the cellular effects of chloroquine.
The team said these findings broaden our understanding of Cam734, the second most common variant of the pfcrt gene in Southeast Asia. The findings identify multiple intracellular processes and multidrug resistance phenotypes impacted by changes in pfcrt and can help inform future malaria treatment efforts.

Plasmodium falciparum
Image from CDC/Mae Melvin
Some mutations that enable drug resistance in the malaria parasite Plasmodium falciparum may also help it grow, according to a study published in PLOS Pathogens.
Some strains of P falciparum have evolved to become resistant to antimalarial drugs, including chloroquine.
Often, chloroquine resistance mutations hinder P falciparum’s ability to infect the bloodstream and grow.
However, in a previous study, researchers discovered that a uniquely mutated version of the P falciparum gene pfcrt provides drug resistance while avoiding the detrimental impact of growth seen with other mutated pfcrt variants.
In the new study, the same group of researchers—Stanislaw Gabryszewski, of Columbia University Medical Center in New York, and his colleagues—investigated this version of the pfcrt gene, which is called Cam734 and has been found in certain regions in Southeast Asia.
Using zinc-finger nucleases, the team characterized the individual mutations unique to Cam734 in terms of their effects on drug resistance, metabolism, and growth rates in living parasites.
The researchers found that a mutation called A144F is required for the chloroquine resistance enabled by Cam734, and this mutation also contributes to resistance to the drugs amodiaquine and quinine.
The team identified additional mutations that contribute to resistance to chloroquine and impact the potency of other antimalarials as well.
When the researchers reversed these mutations in living parasites that had the Cam734 allele, growth slowed, indicating that these mutations also enhance infection.
Additional experiments revealed specific effects of Cam734 mutations on several metabolic pathways in P falciparum, including the digestion of human hemoglobin that parasites use to obtain amino acids for protein synthesis.
The researchers also found evidence suggesting that Cam734 helps to maintain an electrochemical gradient that allows the protein encoded by the pfcrt gene to thwart the cellular effects of chloroquine.
The team said these findings broaden our understanding of Cam734, the second most common variant of the pfcrt gene in Southeast Asia. The findings identify multiple intracellular processes and multidrug resistance phenotypes impacted by changes in pfcrt and can help inform future malaria treatment efforts.

Plasmodium falciparum
Image from CDC/Mae Melvin
Some mutations that enable drug resistance in the malaria parasite Plasmodium falciparum may also help it grow, according to a study published in PLOS Pathogens.
Some strains of P falciparum have evolved to become resistant to antimalarial drugs, including chloroquine.
Often, chloroquine resistance mutations hinder P falciparum’s ability to infect the bloodstream and grow.
However, in a previous study, researchers discovered that a uniquely mutated version of the P falciparum gene pfcrt provides drug resistance while avoiding the detrimental impact of growth seen with other mutated pfcrt variants.
In the new study, the same group of researchers—Stanislaw Gabryszewski, of Columbia University Medical Center in New York, and his colleagues—investigated this version of the pfcrt gene, which is called Cam734 and has been found in certain regions in Southeast Asia.
Using zinc-finger nucleases, the team characterized the individual mutations unique to Cam734 in terms of their effects on drug resistance, metabolism, and growth rates in living parasites.
The researchers found that a mutation called A144F is required for the chloroquine resistance enabled by Cam734, and this mutation also contributes to resistance to the drugs amodiaquine and quinine.
The team identified additional mutations that contribute to resistance to chloroquine and impact the potency of other antimalarials as well.
When the researchers reversed these mutations in living parasites that had the Cam734 allele, growth slowed, indicating that these mutations also enhance infection.
Additional experiments revealed specific effects of Cam734 mutations on several metabolic pathways in P falciparum, including the digestion of human hemoglobin that parasites use to obtain amino acids for protein synthesis.
The researchers also found evidence suggesting that Cam734 helps to maintain an electrochemical gradient that allows the protein encoded by the pfcrt gene to thwart the cellular effects of chloroquine.
The team said these findings broaden our understanding of Cam734, the second most common variant of the pfcrt gene in Southeast Asia. The findings identify multiple intracellular processes and multidrug resistance phenotypes impacted by changes in pfcrt and can help inform future malaria treatment efforts.
CHMP recommends drug for hemophilia A
The European Medicines Agency’s Committee for Medicinal Products for Human Use (CHMP) has recommended marketing authorization for lonoctocog alfa (Afstyla), a recombinant factor VIII (FVIII) single-chain therapy.
Lonoctocog alfa is intended for the treatment and prophylaxis of bleeding in hemophilia A patients of all ages.
The CHMP’s recommendation will be reviewed by the European Commission, which is expected to make a decision in the next few months.
Lonoctocog alfa is designed to provide lasting protection from bleeds with 2- to 3-times weekly dosing. The product uses a covalent bond that forms one structural entity, a single polypeptide-chain, to improve the stability of FVIII and provide longer-lasting FVIII activity.
Lonoctocog alfa is being developed by CSL Behring GmbH.
Clinical trials
The CHMP’s positive opinion of lonoctocog alfa is based on results from the AFFINITY clinical development program, which includes a trial of children (n=84) and a trial of adolescents and adults (n=175).
Among patients who received lonoctocog alfa prophylactically, the median annualized bleeding rate was 1.14 in the adults and adolescents and 3.69 in children younger than 12.
In all, there were 1195 bleeding events—848 in the adults/adolescents and 347 in the children.
Ninety-four percent of bleeds in adults/adolescents and 96% of bleeds in pediatric patients were effectively controlled with no more than 2 infusions of lonoctocog alfa weekly.
Eighty-one percent of bleeds in adults/adolescents and 86% of bleeds in pediatric patients were controlled by a single infusion.
Researchers assessed safety in 258 patients from both studies. Adverse reactions occurred in 14 patients and included hypersensitivity (n=4), dizziness (n=2), paresthesia (n=1), rash (n=1), erythema (n=1), pruritus (n=1), pyrexia (n=1), injection-site pain (n=1), chills (n=1), and feeling hot (n=1).
One patient withdrew from treatment due to hypersensitivity.
None of the patients developed neutralizing antibodies to FVIII or antibodies to host cell proteins. There were no reports of anaphylaxis or thrombosis.

The European Medicines Agency’s Committee for Medicinal Products for Human Use (CHMP) has recommended marketing authorization for lonoctocog alfa (Afstyla), a recombinant factor VIII (FVIII) single-chain therapy.
Lonoctocog alfa is intended for the treatment and prophylaxis of bleeding in hemophilia A patients of all ages.
The CHMP’s recommendation will be reviewed by the European Commission, which is expected to make a decision in the next few months.
Lonoctocog alfa is designed to provide lasting protection from bleeds with 2- to 3-times weekly dosing. The product uses a covalent bond that forms one structural entity, a single polypeptide-chain, to improve the stability of FVIII and provide longer-lasting FVIII activity.
Lonoctocog alfa is being developed by CSL Behring GmbH.
Clinical trials
The CHMP’s positive opinion of lonoctocog alfa is based on results from the AFFINITY clinical development program, which includes a trial of children (n=84) and a trial of adolescents and adults (n=175).
Among patients who received lonoctocog alfa prophylactically, the median annualized bleeding rate was 1.14 in the adults and adolescents and 3.69 in children younger than 12.
In all, there were 1195 bleeding events—848 in the adults/adolescents and 347 in the children.
Ninety-four percent of bleeds in adults/adolescents and 96% of bleeds in pediatric patients were effectively controlled with no more than 2 infusions of lonoctocog alfa weekly.
Eighty-one percent of bleeds in adults/adolescents and 86% of bleeds in pediatric patients were controlled by a single infusion.
Researchers assessed safety in 258 patients from both studies. Adverse reactions occurred in 14 patients and included hypersensitivity (n=4), dizziness (n=2), paresthesia (n=1), rash (n=1), erythema (n=1), pruritus (n=1), pyrexia (n=1), injection-site pain (n=1), chills (n=1), and feeling hot (n=1).
One patient withdrew from treatment due to hypersensitivity.
None of the patients developed neutralizing antibodies to FVIII or antibodies to host cell proteins. There were no reports of anaphylaxis or thrombosis.

The European Medicines Agency’s Committee for Medicinal Products for Human Use (CHMP) has recommended marketing authorization for lonoctocog alfa (Afstyla), a recombinant factor VIII (FVIII) single-chain therapy.
Lonoctocog alfa is intended for the treatment and prophylaxis of bleeding in hemophilia A patients of all ages.
The CHMP’s recommendation will be reviewed by the European Commission, which is expected to make a decision in the next few months.
Lonoctocog alfa is designed to provide lasting protection from bleeds with 2- to 3-times weekly dosing. The product uses a covalent bond that forms one structural entity, a single polypeptide-chain, to improve the stability of FVIII and provide longer-lasting FVIII activity.
Lonoctocog alfa is being developed by CSL Behring GmbH.
Clinical trials
The CHMP’s positive opinion of lonoctocog alfa is based on results from the AFFINITY clinical development program, which includes a trial of children (n=84) and a trial of adolescents and adults (n=175).
Among patients who received lonoctocog alfa prophylactically, the median annualized bleeding rate was 1.14 in the adults and adolescents and 3.69 in children younger than 12.
In all, there were 1195 bleeding events—848 in the adults/adolescents and 347 in the children.
Ninety-four percent of bleeds in adults/adolescents and 96% of bleeds in pediatric patients were effectively controlled with no more than 2 infusions of lonoctocog alfa weekly.
Eighty-one percent of bleeds in adults/adolescents and 86% of bleeds in pediatric patients were controlled by a single infusion.
Researchers assessed safety in 258 patients from both studies. Adverse reactions occurred in 14 patients and included hypersensitivity (n=4), dizziness (n=2), paresthesia (n=1), rash (n=1), erythema (n=1), pruritus (n=1), pyrexia (n=1), injection-site pain (n=1), chills (n=1), and feeling hot (n=1).
One patient withdrew from treatment due to hypersensitivity.
None of the patients developed neutralizing antibodies to FVIII or antibodies to host cell proteins. There were no reports of anaphylaxis or thrombosis.