User login
Appalachia has higher cancer incidence than rest of US
receiving chemotherapy
Photo by Rhoda Baer
New research suggests that people living in the Appalachian region of the US are more likely to develop cancer than people in the rest of the country.
The study showed that Appalachians had a significantly higher incidence of cancer overall and higher rates of many solid tumor malignancies.
However, lymphoma rates were similar between Appalachians and non-Appalachians, and Appalachians had a significantly lower rate of myeloma.
This research was published in Cancer Epidemiology, Biomarkers & Prevention.
“The Appalachian region, which extends from parts of New York to Mississippi, spans 420 counties in 13 US states, and about 25 million people reside in this area,” said study author Reda Wilson, MPH, of the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia.
“This region is primarily made up of rural areas, with persistent poverty levels that are at least 20%, which is higher than the national average.”
In 2007, the CDC’s National Program of Cancer Registries (NPCR) published a comprehensive evaluation of cancer incidence rates in Appalachia between 2001 and 2003.
The data showed higher cancer rates in Appalachia than in the rest of the US. However, this publication had some shortcomings, including data that were not available for analysis.
“The current analyses reported here were performed to update the earlier evaluation by expanding the diagnosis years from 2004 to 2011 and including data on 100% of the Appalachian and non-Appalachian populations,” Wilson said.
For this study, Wilson and her colleagues used data from the NPCR and the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program. Together, NPCR and SEER cover 100% of the US population.
The researchers analyzed the Appalachian population by region (north, central, and south Appalachia), gender, race (black and white only), and Appalachian Regional Commission-designated economic status (distressed, at-risk, transitional, competitive, and attainment). And the team compared these data with data on the non-Appalachian population.
The results showed that cancer incidence rates (IRs) were elevated among Appalachians regardless of how they were categorized. The IRs were per 100,000 people, age-adjusted to the 2000 US standard population.
The IR for all cancers was 565.8 for males in Appalachia and 543.0 for non-Appalachian males (P<0.05). And the cancer IRs for females were 428.7 in Appalachia and 418.2 outside the region (P<0.05).
There was no significant difference between the regions in IRs for lymphomas. The Hodgkin lymphoma IRs were 3.1 in Appalachian males, 3.2 in non-Appalachian males, and 2.5 for females in both regions.
The non-Hodgkin lymphoma IRs were 23.3 in Appalachian males, 23.4 in non-Appalachian males, 16.4 in Appalachian females, and 16.3 in non-Appalachian females.
Myeloma IRs were significantly lower in Appalachia (P<0.05). The myeloma IRs were 7.3 in Appalachian males, 7.5 in non-Appalachian males, 4.7 in Appalachian females, and 4.9 in non-Appalachian females.
There was no significant difference in leukemia IRs among males, but females in Appalachia had a significantly higher leukemia IR (P<0.05). The leukemia IRs were 16.9 in Appalachian males, 16.7 in non-Appalachian males, 10.4 in Appalachian females, and 10.2 in non-Appalachian females.
“Appalachia continues to have higher cancer incidence rates than the rest of the country,” Wilson said. “But a promising finding is that we’re seeing the gap narrow in the incidence rates between Appalachia and non-Appalachia since the 2007 analysis, with the exception of cancers of the oral cavity and pharynx, larynx, lung and bronchus, and thyroid.”
“This study helps identify types of cancer in the Appalachian region that could be reduced through more evidence-based screening and detection. Our study also emphasizes the importance of lifestyle changes needed to prevent and reduce cancer burden.”
The researchers noted that this study did not differentiate urban versus rural areas within each county, and data on screening and risk factors were based on self-reported responses.
Furthermore, cancer IRs were calculated for all ages combined and were not evaluated by age groups. Future analyses will be targeted toward capturing these finer details, Wilson said. ![]()

receiving chemotherapy
Photo by Rhoda Baer
New research suggests that people living in the Appalachian region of the US are more likely to develop cancer than people in the rest of the country.
The study showed that Appalachians had a significantly higher incidence of cancer overall and higher rates of many solid tumor malignancies.
However, lymphoma rates were similar between Appalachians and non-Appalachians, and Appalachians had a significantly lower rate of myeloma.
This research was published in Cancer Epidemiology, Biomarkers & Prevention.
“The Appalachian region, which extends from parts of New York to Mississippi, spans 420 counties in 13 US states, and about 25 million people reside in this area,” said study author Reda Wilson, MPH, of the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia.
“This region is primarily made up of rural areas, with persistent poverty levels that are at least 20%, which is higher than the national average.”
In 2007, the CDC’s National Program of Cancer Registries (NPCR) published a comprehensive evaluation of cancer incidence rates in Appalachia between 2001 and 2003.
The data showed higher cancer rates in Appalachia than in the rest of the US. However, this publication had some shortcomings, including data that were not available for analysis.
“The current analyses reported here were performed to update the earlier evaluation by expanding the diagnosis years from 2004 to 2011 and including data on 100% of the Appalachian and non-Appalachian populations,” Wilson said.
For this study, Wilson and her colleagues used data from the NPCR and the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program. Together, NPCR and SEER cover 100% of the US population.
The researchers analyzed the Appalachian population by region (north, central, and south Appalachia), gender, race (black and white only), and Appalachian Regional Commission-designated economic status (distressed, at-risk, transitional, competitive, and attainment). And the team compared these data with data on the non-Appalachian population.
The results showed that cancer incidence rates (IRs) were elevated among Appalachians regardless of how they were categorized. The IRs were per 100,000 people, age-adjusted to the 2000 US standard population.
The IR for all cancers was 565.8 for males in Appalachia and 543.0 for non-Appalachian males (P<0.05). And the cancer IRs for females were 428.7 in Appalachia and 418.2 outside the region (P<0.05).
There was no significant difference between the regions in IRs for lymphomas. The Hodgkin lymphoma IRs were 3.1 in Appalachian males, 3.2 in non-Appalachian males, and 2.5 for females in both regions.
The non-Hodgkin lymphoma IRs were 23.3 in Appalachian males, 23.4 in non-Appalachian males, 16.4 in Appalachian females, and 16.3 in non-Appalachian females.
Myeloma IRs were significantly lower in Appalachia (P<0.05). The myeloma IRs were 7.3 in Appalachian males, 7.5 in non-Appalachian males, 4.7 in Appalachian females, and 4.9 in non-Appalachian females.
There was no significant difference in leukemia IRs among males, but females in Appalachia had a significantly higher leukemia IR (P<0.05). The leukemia IRs were 16.9 in Appalachian males, 16.7 in non-Appalachian males, 10.4 in Appalachian females, and 10.2 in non-Appalachian females.
“Appalachia continues to have higher cancer incidence rates than the rest of the country,” Wilson said. “But a promising finding is that we’re seeing the gap narrow in the incidence rates between Appalachia and non-Appalachia since the 2007 analysis, with the exception of cancers of the oral cavity and pharynx, larynx, lung and bronchus, and thyroid.”
“This study helps identify types of cancer in the Appalachian region that could be reduced through more evidence-based screening and detection. Our study also emphasizes the importance of lifestyle changes needed to prevent and reduce cancer burden.”
The researchers noted that this study did not differentiate urban versus rural areas within each county, and data on screening and risk factors were based on self-reported responses.
Furthermore, cancer IRs were calculated for all ages combined and were not evaluated by age groups. Future analyses will be targeted toward capturing these finer details, Wilson said. ![]()

receiving chemotherapy
Photo by Rhoda Baer
New research suggests that people living in the Appalachian region of the US are more likely to develop cancer than people in the rest of the country.
The study showed that Appalachians had a significantly higher incidence of cancer overall and higher rates of many solid tumor malignancies.
However, lymphoma rates were similar between Appalachians and non-Appalachians, and Appalachians had a significantly lower rate of myeloma.
This research was published in Cancer Epidemiology, Biomarkers & Prevention.
“The Appalachian region, which extends from parts of New York to Mississippi, spans 420 counties in 13 US states, and about 25 million people reside in this area,” said study author Reda Wilson, MPH, of the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia.
“This region is primarily made up of rural areas, with persistent poverty levels that are at least 20%, which is higher than the national average.”
In 2007, the CDC’s National Program of Cancer Registries (NPCR) published a comprehensive evaluation of cancer incidence rates in Appalachia between 2001 and 2003.
The data showed higher cancer rates in Appalachia than in the rest of the US. However, this publication had some shortcomings, including data that were not available for analysis.
“The current analyses reported here were performed to update the earlier evaluation by expanding the diagnosis years from 2004 to 2011 and including data on 100% of the Appalachian and non-Appalachian populations,” Wilson said.
For this study, Wilson and her colleagues used data from the NPCR and the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program. Together, NPCR and SEER cover 100% of the US population.
The researchers analyzed the Appalachian population by region (north, central, and south Appalachia), gender, race (black and white only), and Appalachian Regional Commission-designated economic status (distressed, at-risk, transitional, competitive, and attainment). And the team compared these data with data on the non-Appalachian population.
The results showed that cancer incidence rates (IRs) were elevated among Appalachians regardless of how they were categorized. The IRs were per 100,000 people, age-adjusted to the 2000 US standard population.
The IR for all cancers was 565.8 for males in Appalachia and 543.0 for non-Appalachian males (P<0.05). And the cancer IRs for females were 428.7 in Appalachia and 418.2 outside the region (P<0.05).
There was no significant difference between the regions in IRs for lymphomas. The Hodgkin lymphoma IRs were 3.1 in Appalachian males, 3.2 in non-Appalachian males, and 2.5 for females in both regions.
The non-Hodgkin lymphoma IRs were 23.3 in Appalachian males, 23.4 in non-Appalachian males, 16.4 in Appalachian females, and 16.3 in non-Appalachian females.
Myeloma IRs were significantly lower in Appalachia (P<0.05). The myeloma IRs were 7.3 in Appalachian males, 7.5 in non-Appalachian males, 4.7 in Appalachian females, and 4.9 in non-Appalachian females.
There was no significant difference in leukemia IRs among males, but females in Appalachia had a significantly higher leukemia IR (P<0.05). The leukemia IRs were 16.9 in Appalachian males, 16.7 in non-Appalachian males, 10.4 in Appalachian females, and 10.2 in non-Appalachian females.
“Appalachia continues to have higher cancer incidence rates than the rest of the country,” Wilson said. “But a promising finding is that we’re seeing the gap narrow in the incidence rates between Appalachia and non-Appalachia since the 2007 analysis, with the exception of cancers of the oral cavity and pharynx, larynx, lung and bronchus, and thyroid.”
“This study helps identify types of cancer in the Appalachian region that could be reduced through more evidence-based screening and detection. Our study also emphasizes the importance of lifestyle changes needed to prevent and reduce cancer burden.”
The researchers noted that this study did not differentiate urban versus rural areas within each county, and data on screening and risk factors were based on self-reported responses.
Furthermore, cancer IRs were calculated for all ages combined and were not evaluated by age groups. Future analyses will be targeted toward capturing these finer details, Wilson said. ![]()
Method makes gene editing more efficient, team says

Image by Spencer Phillips
Researchers say they have made an improvement in CRISPR-Cas9 technology that enables a success rate of 60% when replacing a short stretch of DNA
with another.
They say this technique could be especially useful when trying to repair genetic mutations that cause hereditary diseases, such as sickle cell disease.
The technique allows researchers to patch an abnormal section of DNA with the normal sequence and potentially correct the defect.
“The exciting thing about CRISPR-Cas9 is the promise of fixing genes in place in our genome, but the efficiency for that can be very low,” said Jacob Corn, PhD, of the University of California, Berkeley.
“If you think of gene editing as a word processor, we know how to cut, but we need a more efficient way to paste and glue a new piece of DNA where we make the cut.”
Dr Corn and his colleagues described their more efficient technique in Nature Biotechnology.
“In cases where you want to change very small regions of DNA, up to 30 base pairs, this technique would be extremely effective,” said study author Christopher Richardson, PhD, of the University of California, Berkeley.
Dr Richardson invented the new approach after finding that the Cas9 protein, which does the actual DNA cutting, remains attached to the chromosome for up to 6 hours, long after it has sliced through the double-stranded DNA.
Dr Richardson looked closely at the Cas9 protein bound to the 2 strands of DNA and discovered that while the protein hangs onto 3 of the cut ends, 1 of the ends remains free.
When Cas9 cuts DNA, repair systems in the cell can grab a piece of complementary DNA, called a template, to repair the cut. Researchers can add templates containing changes that alter existing sequences in the genome.
Dr Richardson reasoned that bringing the substitute template directly to the site of the cut would improve the patching efficiency. So he constructed a piece of DNA that matches the free DNA end and carries the genetic sequence to be inserted at the other end.
The technique allowed successful repair of a mutation with up to 60% efficiency.
“Our data indicate that Cas9 breaks could be different, at a molecular level, from breaks generated by other targeted nucleases, such as TALENS and zinc-finger nucleases, which suggests that strategies like the ones we are using can give you more efficient repair of Cas9 breaks,” Dr Richardson said.
The researchers also showed that variants of the Cas9 protein that bind DNA but do not cut can also paste a new DNA sequence at the binding site, possibly by forming a “bubble” structure on the target DNA that also acts to attract the repair template.
Gene editing using Cas9 without genome cutting could be safer than typical gene editing by removing the danger of off-target cutting in the genome, Dr Corn said. ![]()

Image by Spencer Phillips
Researchers say they have made an improvement in CRISPR-Cas9 technology that enables a success rate of 60% when replacing a short stretch of DNA
with another.
They say this technique could be especially useful when trying to repair genetic mutations that cause hereditary diseases, such as sickle cell disease.
The technique allows researchers to patch an abnormal section of DNA with the normal sequence and potentially correct the defect.
“The exciting thing about CRISPR-Cas9 is the promise of fixing genes in place in our genome, but the efficiency for that can be very low,” said Jacob Corn, PhD, of the University of California, Berkeley.
“If you think of gene editing as a word processor, we know how to cut, but we need a more efficient way to paste and glue a new piece of DNA where we make the cut.”
Dr Corn and his colleagues described their more efficient technique in Nature Biotechnology.
“In cases where you want to change very small regions of DNA, up to 30 base pairs, this technique would be extremely effective,” said study author Christopher Richardson, PhD, of the University of California, Berkeley.
Dr Richardson invented the new approach after finding that the Cas9 protein, which does the actual DNA cutting, remains attached to the chromosome for up to 6 hours, long after it has sliced through the double-stranded DNA.
Dr Richardson looked closely at the Cas9 protein bound to the 2 strands of DNA and discovered that while the protein hangs onto 3 of the cut ends, 1 of the ends remains free.
When Cas9 cuts DNA, repair systems in the cell can grab a piece of complementary DNA, called a template, to repair the cut. Researchers can add templates containing changes that alter existing sequences in the genome.
Dr Richardson reasoned that bringing the substitute template directly to the site of the cut would improve the patching efficiency. So he constructed a piece of DNA that matches the free DNA end and carries the genetic sequence to be inserted at the other end.
The technique allowed successful repair of a mutation with up to 60% efficiency.
“Our data indicate that Cas9 breaks could be different, at a molecular level, from breaks generated by other targeted nucleases, such as TALENS and zinc-finger nucleases, which suggests that strategies like the ones we are using can give you more efficient repair of Cas9 breaks,” Dr Richardson said.
The researchers also showed that variants of the Cas9 protein that bind DNA but do not cut can also paste a new DNA sequence at the binding site, possibly by forming a “bubble” structure on the target DNA that also acts to attract the repair template.
Gene editing using Cas9 without genome cutting could be safer than typical gene editing by removing the danger of off-target cutting in the genome, Dr Corn said. ![]()

Image by Spencer Phillips
Researchers say they have made an improvement in CRISPR-Cas9 technology that enables a success rate of 60% when replacing a short stretch of DNA
with another.
They say this technique could be especially useful when trying to repair genetic mutations that cause hereditary diseases, such as sickle cell disease.
The technique allows researchers to patch an abnormal section of DNA with the normal sequence and potentially correct the defect.
“The exciting thing about CRISPR-Cas9 is the promise of fixing genes in place in our genome, but the efficiency for that can be very low,” said Jacob Corn, PhD, of the University of California, Berkeley.
“If you think of gene editing as a word processor, we know how to cut, but we need a more efficient way to paste and glue a new piece of DNA where we make the cut.”
Dr Corn and his colleagues described their more efficient technique in Nature Biotechnology.
“In cases where you want to change very small regions of DNA, up to 30 base pairs, this technique would be extremely effective,” said study author Christopher Richardson, PhD, of the University of California, Berkeley.
Dr Richardson invented the new approach after finding that the Cas9 protein, which does the actual DNA cutting, remains attached to the chromosome for up to 6 hours, long after it has sliced through the double-stranded DNA.
Dr Richardson looked closely at the Cas9 protein bound to the 2 strands of DNA and discovered that while the protein hangs onto 3 of the cut ends, 1 of the ends remains free.
When Cas9 cuts DNA, repair systems in the cell can grab a piece of complementary DNA, called a template, to repair the cut. Researchers can add templates containing changes that alter existing sequences in the genome.
Dr Richardson reasoned that bringing the substitute template directly to the site of the cut would improve the patching efficiency. So he constructed a piece of DNA that matches the free DNA end and carries the genetic sequence to be inserted at the other end.
The technique allowed successful repair of a mutation with up to 60% efficiency.
“Our data indicate that Cas9 breaks could be different, at a molecular level, from breaks generated by other targeted nucleases, such as TALENS and zinc-finger nucleases, which suggests that strategies like the ones we are using can give you more efficient repair of Cas9 breaks,” Dr Richardson said.
The researchers also showed that variants of the Cas9 protein that bind DNA but do not cut can also paste a new DNA sequence at the binding site, possibly by forming a “bubble” structure on the target DNA that also acts to attract the repair template.
Gene editing using Cas9 without genome cutting could be safer than typical gene editing by removing the danger of off-target cutting in the genome, Dr Corn said. ![]()
Generic drugs often out of reach, experts say

Photo courtesy of the CDC
An article published in Blood suggests pharmaceutical companies use several strategies to keep affordable generic drugs from the US market.
“The timely availability of affordable generic drugs is the difference between life or death for patients with cancer and other diseases who cannot afford brand-name pharmaceuticals, the majority of which are priced at monopoly levels and protected by 20-year patents,” said lead author Hagop Kantarjian, MD, of The University of Texas MD Anderson Cancer Center in Houston.
“Unfortunately, these sorely needed generics are increasingly out of reach. As we sought to understand what keeps these affordable drugs from the market, we identified several specific strategies that pharmaceutical companies use to extend their patents and eliminate competition.”
Dr Kantarjian and his colleagues assert that pharmaceutical companies use a variety of strategies to delay, prevent, and suppress the timely availability of affordable generic drugs.
Among them, the authors detail “pay-for-delay,” in which the company that owns the patent pays a generic company to delay entry into the market. The Federal Trade Commission estimates that the pay-for-delay settlements cost taxpayers, insurance companies, and consumers approximately $3.5 billion per year.
In other cases detailed in the article, the patent-holder deters competition by creating its own version of drugs at generic prices.
While this practice may reduce costs for consumers by 4% to 8% in the short-term, the authors suggest that companies often use the authorized generics as a bargaining chip in “pay-for-delay” deals, pledging not to release their own drugs in return for the true generic company promising to delay market entry.
Other strategies the authors discuss include investing heavily in advertising the brand-name drug (often spending more on marketing than on research and development) and lobbying for laws that prevent patients from importing cheaper generics from other countries, which the authors write can cost as little as 20% to 50% of US prices.
The authors also say some drug companies buy out competitors and then increase the price of a newly acquired generic drug by several fold overnight.
In addition, the authors describe a strategy they call “product hopping,” which involves switching the market for a drug to a reformulated “new and improved” version with a slightly different tablet or capsule dose that offers no therapeutic advantage over the original but has a later-expiring patent.
The company then heavily advertises the new brand-name drug in an effort to convince patients and physicians to switch.
As a result, when the generic version of the original becomes available, pharmacists cannot substitute it for the new branded version because state laws allow substitution only if certain characteristics, such as dosing, remain the same.
In recognition of the harm and expense the authors suggest these strategies impart on both patients and the economy, they propose several solutions that would support timely access to affordable generic drugs.
These include allowing Medicare to negotiate drug prices, monitoring and penalizing pay-for-delay deals, allowing transportation of pharmaceuticals across borders for individual use, and challenging weak patents.
“Each day, in my clinic, I see leukemia patients who are harmed because they cannot afford their treatment, some risking death because they cannot pay for the medicine keeping them alive,” Dr Kantarjian said.
“Overall, these strategies demonstrate that the trend of high brand-name drug prices has recently infected generic drugs, as companies value profit at the expense of long-term utility to society. We must be vigilant in recognizing these strategies and advocating for solutions that will allow companies to accomplish their dual mission: make reasonable profits and help save and/or improve patients’ lives.” ![]()

Photo courtesy of the CDC
An article published in Blood suggests pharmaceutical companies use several strategies to keep affordable generic drugs from the US market.
“The timely availability of affordable generic drugs is the difference between life or death for patients with cancer and other diseases who cannot afford brand-name pharmaceuticals, the majority of which are priced at monopoly levels and protected by 20-year patents,” said lead author Hagop Kantarjian, MD, of The University of Texas MD Anderson Cancer Center in Houston.
“Unfortunately, these sorely needed generics are increasingly out of reach. As we sought to understand what keeps these affordable drugs from the market, we identified several specific strategies that pharmaceutical companies use to extend their patents and eliminate competition.”
Dr Kantarjian and his colleagues assert that pharmaceutical companies use a variety of strategies to delay, prevent, and suppress the timely availability of affordable generic drugs.
Among them, the authors detail “pay-for-delay,” in which the company that owns the patent pays a generic company to delay entry into the market. The Federal Trade Commission estimates that the pay-for-delay settlements cost taxpayers, insurance companies, and consumers approximately $3.5 billion per year.
In other cases detailed in the article, the patent-holder deters competition by creating its own version of drugs at generic prices.
While this practice may reduce costs for consumers by 4% to 8% in the short-term, the authors suggest that companies often use the authorized generics as a bargaining chip in “pay-for-delay” deals, pledging not to release their own drugs in return for the true generic company promising to delay market entry.
Other strategies the authors discuss include investing heavily in advertising the brand-name drug (often spending more on marketing than on research and development) and lobbying for laws that prevent patients from importing cheaper generics from other countries, which the authors write can cost as little as 20% to 50% of US prices.
The authors also say some drug companies buy out competitors and then increase the price of a newly acquired generic drug by several fold overnight.
In addition, the authors describe a strategy they call “product hopping,” which involves switching the market for a drug to a reformulated “new and improved” version with a slightly different tablet or capsule dose that offers no therapeutic advantage over the original but has a later-expiring patent.
The company then heavily advertises the new brand-name drug in an effort to convince patients and physicians to switch.
As a result, when the generic version of the original becomes available, pharmacists cannot substitute it for the new branded version because state laws allow substitution only if certain characteristics, such as dosing, remain the same.
In recognition of the harm and expense the authors suggest these strategies impart on both patients and the economy, they propose several solutions that would support timely access to affordable generic drugs.
These include allowing Medicare to negotiate drug prices, monitoring and penalizing pay-for-delay deals, allowing transportation of pharmaceuticals across borders for individual use, and challenging weak patents.
“Each day, in my clinic, I see leukemia patients who are harmed because they cannot afford their treatment, some risking death because they cannot pay for the medicine keeping them alive,” Dr Kantarjian said.
“Overall, these strategies demonstrate that the trend of high brand-name drug prices has recently infected generic drugs, as companies value profit at the expense of long-term utility to society. We must be vigilant in recognizing these strategies and advocating for solutions that will allow companies to accomplish their dual mission: make reasonable profits and help save and/or improve patients’ lives.” ![]()

Photo courtesy of the CDC
An article published in Blood suggests pharmaceutical companies use several strategies to keep affordable generic drugs from the US market.
“The timely availability of affordable generic drugs is the difference between life or death for patients with cancer and other diseases who cannot afford brand-name pharmaceuticals, the majority of which are priced at monopoly levels and protected by 20-year patents,” said lead author Hagop Kantarjian, MD, of The University of Texas MD Anderson Cancer Center in Houston.
“Unfortunately, these sorely needed generics are increasingly out of reach. As we sought to understand what keeps these affordable drugs from the market, we identified several specific strategies that pharmaceutical companies use to extend their patents and eliminate competition.”
Dr Kantarjian and his colleagues assert that pharmaceutical companies use a variety of strategies to delay, prevent, and suppress the timely availability of affordable generic drugs.
Among them, the authors detail “pay-for-delay,” in which the company that owns the patent pays a generic company to delay entry into the market. The Federal Trade Commission estimates that the pay-for-delay settlements cost taxpayers, insurance companies, and consumers approximately $3.5 billion per year.
In other cases detailed in the article, the patent-holder deters competition by creating its own version of drugs at generic prices.
While this practice may reduce costs for consumers by 4% to 8% in the short-term, the authors suggest that companies often use the authorized generics as a bargaining chip in “pay-for-delay” deals, pledging not to release their own drugs in return for the true generic company promising to delay market entry.
Other strategies the authors discuss include investing heavily in advertising the brand-name drug (often spending more on marketing than on research and development) and lobbying for laws that prevent patients from importing cheaper generics from other countries, which the authors write can cost as little as 20% to 50% of US prices.
The authors also say some drug companies buy out competitors and then increase the price of a newly acquired generic drug by several fold overnight.
In addition, the authors describe a strategy they call “product hopping,” which involves switching the market for a drug to a reformulated “new and improved” version with a slightly different tablet or capsule dose that offers no therapeutic advantage over the original but has a later-expiring patent.
The company then heavily advertises the new brand-name drug in an effort to convince patients and physicians to switch.
As a result, when the generic version of the original becomes available, pharmacists cannot substitute it for the new branded version because state laws allow substitution only if certain characteristics, such as dosing, remain the same.
In recognition of the harm and expense the authors suggest these strategies impart on both patients and the economy, they propose several solutions that would support timely access to affordable generic drugs.
These include allowing Medicare to negotiate drug prices, monitoring and penalizing pay-for-delay deals, allowing transportation of pharmaceuticals across borders for individual use, and challenging weak patents.
“Each day, in my clinic, I see leukemia patients who are harmed because they cannot afford their treatment, some risking death because they cannot pay for the medicine keeping them alive,” Dr Kantarjian said.
“Overall, these strategies demonstrate that the trend of high brand-name drug prices has recently infected generic drugs, as companies value profit at the expense of long-term utility to society. We must be vigilant in recognizing these strategies and advocating for solutions that will allow companies to accomplish their dual mission: make reasonable profits and help save and/or improve patients’ lives.” ![]()
Study reveals how Tregs protect themselves
Image by Kathryn Iacono
Researchers say they have discovered how regulatory T cells (Tregs) remain intact and functional during activation.
The team found that once Tregs are activated, they are protected by autophagy, which maintains metabolic balance.
Hongbo Chi, PhD, of St. Jude Children’s Hospital in Memphis, Tennessee, and his colleagues described this discovery in Nature Immunology.
Until this study, no one knew how Tregs maintained themselves when activated.
“Regulatory T cells are very specialized cells that require activation to perform their function in curtailing undesirable immune responses,” Dr Chi explained. “But this activation is a double-edged sword, in that this very activation can destabilize them. They need to modulate this activation, or they will lose their stability and many of them will die. That could damage immune function.”
Dr Chi and his colleagues performed imaging studies in activated Tregs and found that autophagy was functional in the cells.
In experiments with mice, the researchers deleted Atg7 or Atg5, genes whose functions are necessary for autophagy in Tregs.
The mice showed key characteristics of Treg malfunction, including inflammatory and autoimmune disorders. The mice also more readily cleared tumors from their bodies, due to activated immune systems.
Dr Chi said that eliminating autophagy also affected the fate of Tregs.
“Once those T cells lack autophagy activity, they tend to undergo excessive cell death,” he said. “But even for the remaining surviving cells, they tend to be overly activated and lose their identity because they start to behave like non-regulatory T cells. That is why loss of autophagy in regulatory T cells produces a 2-fold effect on both survival and stability.”
Detailed analysis also revealed how the elimination of autophagy affected the basic energy-producing metabolic pathways of Tregs, compromising their function.
Dr Chi said this new understanding of autophagy’s role in Tregs could enable a 2-fold approach to immune therapy for cancers. Namely, by strengthening tumor-associated immune responses, targeting Treg autophagy could act in synergy with strategies that block autophagy in tumor cells.
In this study, the researchers used a transplanted colon cancer cell line. In further studies, they plan to explore the role of autophagy in immune reactions toward other tumor cell types to determine whether such therapies might be effective in a broad range of cancers.
The team also hopes to gain a better understanding of the detailed biochemical mechanisms regulating how autophagy connects to the cell’s metabolic pathways. ![]()

Image by Kathryn Iacono
Researchers say they have discovered how regulatory T cells (Tregs) remain intact and functional during activation.
The team found that once Tregs are activated, they are protected by autophagy, which maintains metabolic balance.
Hongbo Chi, PhD, of St. Jude Children’s Hospital in Memphis, Tennessee, and his colleagues described this discovery in Nature Immunology.
Until this study, no one knew how Tregs maintained themselves when activated.
“Regulatory T cells are very specialized cells that require activation to perform their function in curtailing undesirable immune responses,” Dr Chi explained. “But this activation is a double-edged sword, in that this very activation can destabilize them. They need to modulate this activation, or they will lose their stability and many of them will die. That could damage immune function.”
Dr Chi and his colleagues performed imaging studies in activated Tregs and found that autophagy was functional in the cells.
In experiments with mice, the researchers deleted Atg7 or Atg5, genes whose functions are necessary for autophagy in Tregs.
The mice showed key characteristics of Treg malfunction, including inflammatory and autoimmune disorders. The mice also more readily cleared tumors from their bodies, due to activated immune systems.
Dr Chi said that eliminating autophagy also affected the fate of Tregs.
“Once those T cells lack autophagy activity, they tend to undergo excessive cell death,” he said. “But even for the remaining surviving cells, they tend to be overly activated and lose their identity because they start to behave like non-regulatory T cells. That is why loss of autophagy in regulatory T cells produces a 2-fold effect on both survival and stability.”
Detailed analysis also revealed how the elimination of autophagy affected the basic energy-producing metabolic pathways of Tregs, compromising their function.
Dr Chi said this new understanding of autophagy’s role in Tregs could enable a 2-fold approach to immune therapy for cancers. Namely, by strengthening tumor-associated immune responses, targeting Treg autophagy could act in synergy with strategies that block autophagy in tumor cells.
In this study, the researchers used a transplanted colon cancer cell line. In further studies, they plan to explore the role of autophagy in immune reactions toward other tumor cell types to determine whether such therapies might be effective in a broad range of cancers.
The team also hopes to gain a better understanding of the detailed biochemical mechanisms regulating how autophagy connects to the cell’s metabolic pathways. ![]()

Image by Kathryn Iacono
Researchers say they have discovered how regulatory T cells (Tregs) remain intact and functional during activation.
The team found that once Tregs are activated, they are protected by autophagy, which maintains metabolic balance.
Hongbo Chi, PhD, of St. Jude Children’s Hospital in Memphis, Tennessee, and his colleagues described this discovery in Nature Immunology.
Until this study, no one knew how Tregs maintained themselves when activated.
“Regulatory T cells are very specialized cells that require activation to perform their function in curtailing undesirable immune responses,” Dr Chi explained. “But this activation is a double-edged sword, in that this very activation can destabilize them. They need to modulate this activation, or they will lose their stability and many of them will die. That could damage immune function.”
Dr Chi and his colleagues performed imaging studies in activated Tregs and found that autophagy was functional in the cells.
In experiments with mice, the researchers deleted Atg7 or Atg5, genes whose functions are necessary for autophagy in Tregs.
The mice showed key characteristics of Treg malfunction, including inflammatory and autoimmune disorders. The mice also more readily cleared tumors from their bodies, due to activated immune systems.
Dr Chi said that eliminating autophagy also affected the fate of Tregs.
“Once those T cells lack autophagy activity, they tend to undergo excessive cell death,” he said. “But even for the remaining surviving cells, they tend to be overly activated and lose their identity because they start to behave like non-regulatory T cells. That is why loss of autophagy in regulatory T cells produces a 2-fold effect on both survival and stability.”
Detailed analysis also revealed how the elimination of autophagy affected the basic energy-producing metabolic pathways of Tregs, compromising their function.
Dr Chi said this new understanding of autophagy’s role in Tregs could enable a 2-fold approach to immune therapy for cancers. Namely, by strengthening tumor-associated immune responses, targeting Treg autophagy could act in synergy with strategies that block autophagy in tumor cells.
In this study, the researchers used a transplanted colon cancer cell line. In further studies, they plan to explore the role of autophagy in immune reactions toward other tumor cell types to determine whether such therapies might be effective in a broad range of cancers.
The team also hopes to gain a better understanding of the detailed biochemical mechanisms regulating how autophagy connects to the cell’s metabolic pathways. ![]()
Malaria test granted CE mark
Image by Peter H. Seeberger
A malaria test called illumigene® Malaria has received the CE mark, which suggests it meets European health and safety standards.
illumigene Malaria is a molecular test that involves the use of loop-mediated isothermal amplification (LAMP) technology.
According to the test’s developers, it provides results in under an hour, doesn’t require a high level technical expertise, and is up to 80,000 times more sensitive than conventional malaria tests.
illumigene Malaria was developed by Meridian Bioscience, Inc., with technical assistance from the US Centers for Disease Control and Prevention (CDC) and Cheikh Anta Diop University in Dakar, Senegal.
“illumigene Malaria has the potential to change current practices,” said Daouda NDIAYE, PharmD, PhD, of Cheikh Anta Diop University.
“Faster and more accurate diagnosis is vital in the fight against malaria. Earlier diagnosis enables the correct treatment to be prescribed, which leads to better clinical outcomes for the person with malaria and keeps malaria treatments for the right people.”
“Because of submicroscopic parasitemia carriage among the populations, a robust, sensitive, and field-community-deployable screening tool is needed to track the malaria reservoir in pre-elimination regions. illumigene Malaria shows this capacity.”
illumigene Malaria uses LAMP technology to amplify DNA and detect the presence of the malaria parasite.
LAMP technology is isothermal and can be used at room temperature without the need to heat reagents or the material being tested, unlike the rapid diagnostic tests currently used in malaria, which use polymerase chain reaction technology. illumigene Malaria does not require refrigeration.
Meridian Bioscience has worked with experts at the CDC and the Cheikh Anta Diop University of Dakar during the development of illumigene Malaria and collaborated with these organizations to design clinical trials.
According to Meridian, data from more than 200 patients in Senegal validated the performance of illumigene Malaria. The test demonstrated 100% sensitivity and detected infected patients that were missed by conventional testing methods.
Meridian said illumigene Malaria will be distributed in the European, Middle Eastern, and African regions by Meridian Bioscience Europe and in additional international markets by the company’s global distribution network. ![]()

Image by Peter H. Seeberger
A malaria test called illumigene® Malaria has received the CE mark, which suggests it meets European health and safety standards.
illumigene Malaria is a molecular test that involves the use of loop-mediated isothermal amplification (LAMP) technology.
According to the test’s developers, it provides results in under an hour, doesn’t require a high level technical expertise, and is up to 80,000 times more sensitive than conventional malaria tests.
illumigene Malaria was developed by Meridian Bioscience, Inc., with technical assistance from the US Centers for Disease Control and Prevention (CDC) and Cheikh Anta Diop University in Dakar, Senegal.
“illumigene Malaria has the potential to change current practices,” said Daouda NDIAYE, PharmD, PhD, of Cheikh Anta Diop University.
“Faster and more accurate diagnosis is vital in the fight against malaria. Earlier diagnosis enables the correct treatment to be prescribed, which leads to better clinical outcomes for the person with malaria and keeps malaria treatments for the right people.”
“Because of submicroscopic parasitemia carriage among the populations, a robust, sensitive, and field-community-deployable screening tool is needed to track the malaria reservoir in pre-elimination regions. illumigene Malaria shows this capacity.”
illumigene Malaria uses LAMP technology to amplify DNA and detect the presence of the malaria parasite.
LAMP technology is isothermal and can be used at room temperature without the need to heat reagents or the material being tested, unlike the rapid diagnostic tests currently used in malaria, which use polymerase chain reaction technology. illumigene Malaria does not require refrigeration.
Meridian Bioscience has worked with experts at the CDC and the Cheikh Anta Diop University of Dakar during the development of illumigene Malaria and collaborated with these organizations to design clinical trials.
According to Meridian, data from more than 200 patients in Senegal validated the performance of illumigene Malaria. The test demonstrated 100% sensitivity and detected infected patients that were missed by conventional testing methods.
Meridian said illumigene Malaria will be distributed in the European, Middle Eastern, and African regions by Meridian Bioscience Europe and in additional international markets by the company’s global distribution network. ![]()

Image by Peter H. Seeberger
A malaria test called illumigene® Malaria has received the CE mark, which suggests it meets European health and safety standards.
illumigene Malaria is a molecular test that involves the use of loop-mediated isothermal amplification (LAMP) technology.
According to the test’s developers, it provides results in under an hour, doesn’t require a high level technical expertise, and is up to 80,000 times more sensitive than conventional malaria tests.
illumigene Malaria was developed by Meridian Bioscience, Inc., with technical assistance from the US Centers for Disease Control and Prevention (CDC) and Cheikh Anta Diop University in Dakar, Senegal.
“illumigene Malaria has the potential to change current practices,” said Daouda NDIAYE, PharmD, PhD, of Cheikh Anta Diop University.
“Faster and more accurate diagnosis is vital in the fight against malaria. Earlier diagnosis enables the correct treatment to be prescribed, which leads to better clinical outcomes for the person with malaria and keeps malaria treatments for the right people.”
“Because of submicroscopic parasitemia carriage among the populations, a robust, sensitive, and field-community-deployable screening tool is needed to track the malaria reservoir in pre-elimination regions. illumigene Malaria shows this capacity.”
illumigene Malaria uses LAMP technology to amplify DNA and detect the presence of the malaria parasite.
LAMP technology is isothermal and can be used at room temperature without the need to heat reagents or the material being tested, unlike the rapid diagnostic tests currently used in malaria, which use polymerase chain reaction technology. illumigene Malaria does not require refrigeration.
Meridian Bioscience has worked with experts at the CDC and the Cheikh Anta Diop University of Dakar during the development of illumigene Malaria and collaborated with these organizations to design clinical trials.
According to Meridian, data from more than 200 patients in Senegal validated the performance of illumigene Malaria. The test demonstrated 100% sensitivity and detected infected patients that were missed by conventional testing methods.
Meridian said illumigene Malaria will be distributed in the European, Middle Eastern, and African regions by Meridian Bioscience Europe and in additional international markets by the company’s global distribution network. ![]()
Team uncovers new info on hemangioblasts

Research published in PNAS has shed new light on the mechanism by which hemangioblasts become blood cells.
Hemangioblasts, which give rise to both hematopoietic and endothelial progenitors, have been identified in the embryos of chickens, mice, fish, and humans. It has also become clear that the cells are present in adult organisms.
However, the mechanism by which hemangioblasts differentiate into blood cells and vascular endothelia has remained a mystery in many aspects.
With that in mind, Makoto Kobayashi, PhD, of the University of Tsukuba in Japan, and his colleagues studied hemangioblasts in zebrafish.
The investigators looked for zebrafish mutants with defects in the hemangioblast expression of Gata1, which is not expressed in endothelial progenitors.
This revealed a mutant with downregulation of hematopoietic genes and upregulation of endothelial genes.
The team then identified the gene responsible for this mutant—LSD1. Additional experiments showed that LSD1 silences Etv2, a critical regulator of hemangioblast development.
The investigators said these results indicate that epigenetic silencing of Etv2 by LSD1 may be a significant event required for hemangioblasts to initiate hematopoietic differentiation. ![]()

Research published in PNAS has shed new light on the mechanism by which hemangioblasts become blood cells.
Hemangioblasts, which give rise to both hematopoietic and endothelial progenitors, have been identified in the embryos of chickens, mice, fish, and humans. It has also become clear that the cells are present in adult organisms.
However, the mechanism by which hemangioblasts differentiate into blood cells and vascular endothelia has remained a mystery in many aspects.
With that in mind, Makoto Kobayashi, PhD, of the University of Tsukuba in Japan, and his colleagues studied hemangioblasts in zebrafish.
The investigators looked for zebrafish mutants with defects in the hemangioblast expression of Gata1, which is not expressed in endothelial progenitors.
This revealed a mutant with downregulation of hematopoietic genes and upregulation of endothelial genes.
The team then identified the gene responsible for this mutant—LSD1. Additional experiments showed that LSD1 silences Etv2, a critical regulator of hemangioblast development.
The investigators said these results indicate that epigenetic silencing of Etv2 by LSD1 may be a significant event required for hemangioblasts to initiate hematopoietic differentiation. ![]()

Research published in PNAS has shed new light on the mechanism by which hemangioblasts become blood cells.
Hemangioblasts, which give rise to both hematopoietic and endothelial progenitors, have been identified in the embryos of chickens, mice, fish, and humans. It has also become clear that the cells are present in adult organisms.
However, the mechanism by which hemangioblasts differentiate into blood cells and vascular endothelia has remained a mystery in many aspects.
With that in mind, Makoto Kobayashi, PhD, of the University of Tsukuba in Japan, and his colleagues studied hemangioblasts in zebrafish.
The investigators looked for zebrafish mutants with defects in the hemangioblast expression of Gata1, which is not expressed in endothelial progenitors.
This revealed a mutant with downregulation of hematopoietic genes and upregulation of endothelial genes.
The team then identified the gene responsible for this mutant—LSD1. Additional experiments showed that LSD1 silences Etv2, a critical regulator of hemangioblast development.
The investigators said these results indicate that epigenetic silencing of Etv2 by LSD1 may be a significant event required for hemangioblasts to initiate hematopoietic differentiation. ![]()
How p53 and telomeres stave off cancer
telomeres in green
Image by Claus Azzalin
Research published in The EMBO Journal suggests that p53 has tumor suppressor functions related to telomeres.
The study showed, for the first time, that p53 can suppress accumulated DNA damage at telomeres.
P53 is known to regulate the genome’s integrity. When DNA is damaged, p53 helps activate the transcription of genes that regulate the cell cycle and induce apoptosis.
However, prior studies have shown that p53 can bind at many locations across the genome, including sites that are not responsible for activating these regulatory genes.
Paul Lieberman, PhD, of The Wistar Institute in Philadelphia, Pennsylvania, and his colleagues decided to study these binding sites to see if p53 and telomeres might be more closely related than previous research suggested.
“We believed that p53 may be responsible for a more direct protective effect in telomeres,” Dr Lieberman said.
Using ChIP-sequencing, he and his colleagues identified p53 binding sites in subtelomeres.
They found that when p53 was bound to subtelomeres, the protein was able to suppress the formation of a histone modification called γ-H2AX.
This histone is modified in greater amounts when there is a double-strand break on DNA. If it persists, the break is not repaired, so suppressing its expression means the DNA is being preserved.
Additionally, p53 was able to prevent DNA degradation in telomeres, thereby keeping them intact and allowing them to more properly protect the tips of chromosomes.
“Based on our findings, we propose that the modifications to chromatin made by p53 enhance local DNA repair or protection,” Dr Lieberman said. “This would be yet another tumor suppressor function of p53, thus providing additional framework for just how important this gene is in protecting us from cancer.” ![]()

telomeres in green
Image by Claus Azzalin
Research published in The EMBO Journal suggests that p53 has tumor suppressor functions related to telomeres.
The study showed, for the first time, that p53 can suppress accumulated DNA damage at telomeres.
P53 is known to regulate the genome’s integrity. When DNA is damaged, p53 helps activate the transcription of genes that regulate the cell cycle and induce apoptosis.
However, prior studies have shown that p53 can bind at many locations across the genome, including sites that are not responsible for activating these regulatory genes.
Paul Lieberman, PhD, of The Wistar Institute in Philadelphia, Pennsylvania, and his colleagues decided to study these binding sites to see if p53 and telomeres might be more closely related than previous research suggested.
“We believed that p53 may be responsible for a more direct protective effect in telomeres,” Dr Lieberman said.
Using ChIP-sequencing, he and his colleagues identified p53 binding sites in subtelomeres.
They found that when p53 was bound to subtelomeres, the protein was able to suppress the formation of a histone modification called γ-H2AX.
This histone is modified in greater amounts when there is a double-strand break on DNA. If it persists, the break is not repaired, so suppressing its expression means the DNA is being preserved.
Additionally, p53 was able to prevent DNA degradation in telomeres, thereby keeping them intact and allowing them to more properly protect the tips of chromosomes.
“Based on our findings, we propose that the modifications to chromatin made by p53 enhance local DNA repair or protection,” Dr Lieberman said. “This would be yet another tumor suppressor function of p53, thus providing additional framework for just how important this gene is in protecting us from cancer.” ![]()

telomeres in green
Image by Claus Azzalin
Research published in The EMBO Journal suggests that p53 has tumor suppressor functions related to telomeres.
The study showed, for the first time, that p53 can suppress accumulated DNA damage at telomeres.
P53 is known to regulate the genome’s integrity. When DNA is damaged, p53 helps activate the transcription of genes that regulate the cell cycle and induce apoptosis.
However, prior studies have shown that p53 can bind at many locations across the genome, including sites that are not responsible for activating these regulatory genes.
Paul Lieberman, PhD, of The Wistar Institute in Philadelphia, Pennsylvania, and his colleagues decided to study these binding sites to see if p53 and telomeres might be more closely related than previous research suggested.
“We believed that p53 may be responsible for a more direct protective effect in telomeres,” Dr Lieberman said.
Using ChIP-sequencing, he and his colleagues identified p53 binding sites in subtelomeres.
They found that when p53 was bound to subtelomeres, the protein was able to suppress the formation of a histone modification called γ-H2AX.
This histone is modified in greater amounts when there is a double-strand break on DNA. If it persists, the break is not repaired, so suppressing its expression means the DNA is being preserved.
Additionally, p53 was able to prevent DNA degradation in telomeres, thereby keeping them intact and allowing them to more properly protect the tips of chromosomes.
“Based on our findings, we propose that the modifications to chromatin made by p53 enhance local DNA repair or protection,” Dr Lieberman said. “This would be yet another tumor suppressor function of p53, thus providing additional framework for just how important this gene is in protecting us from cancer.”
Fertility concerns of AYA cancer survivors

patient and her father
Photo by Rhoda Baer
Results of a small study suggest that adolescent and young adult (AYA) cancer survivors have a range of concerns regarding their fertility.
The female cancer survivors studied were more likely than males to describe feeling distressed and overwhelmed about their fertility.
Females also tended to worry about pregnancy-related health risks and cancer recurrence.
However, AYA cancer survivors of both sexes expressed concerns about genetic risk factors and how infertility might impact their future lives.
Catherine Benedict, PhD, of Memorial Sloan Kettering Cancer Center in New York, New York, and her colleagues conducted this research and reported the results in the Journal of Adolescent and Young Adult Oncology.
The researchers assessed fertility concerns in 43 AYA cancer survivors. They were 16 to 24 at the time of the study, had been diagnosed with cancer between ages 14 and 18, and were at least 6 months post-treatment.
The subjects either completed an individual interview (n=26) or participated in 1 of 4 focus groups (n=17).
Before treatment, 5 of the males had banked sperm, but none of the females took steps to preserve their fertility. More males (50%) than females (39%) reported uncertainty about their fertility.
The researchers identified 3 themes when discussing fertility with the study subjects: fertility concerns, emotions raised when discussing fertility, and strategies used to manage fertility concerns.
Concerns
Some subjects expressed concerns about how potential infertility could affect dating and relationships with partners (8% of males and 20% of females).
Some subjects were concerned about the health risks associated with having children—both risks to the subjects themselves and to any potential children (39% of males and 30% of females).
And some subjects were concerned about how potential infertility would affect their lives going forward (31% of males and 20% of females).
Emotions
When it came to emotions associated with fertility discussions, some subjects said they felt distressed and overwhelmed (23% of males and 30% of females).
However, some subjects said they weren’t concerned about their fertility (31% of males and 15% of females) or they felt hopeful despite the risk of infertility (8% of males and 20% of females).
Managing concerns
The subjects also mentioned a few strategies for managing fertility concerns. Some said they had accepted infertility (23% of males and 15% of females).
Some subjects said they weren’t going to worry about their fertility until they were older and actually wanted to have children (23% of males and 20% of females).
And some subjects said they would rely on assisted reproductive technology if necessary (31% of males and 20% of females).
Dr Benedict and her colleagues said this study suggests AYA cancer survivors may have a number of reproductive concerns and fertility-related distress, which may affect other areas of psychosocial functioning.
So future research should explore how to best incorporate fertility-related informational and support services more fully into survivorship care.

patient and her father
Photo by Rhoda Baer
Results of a small study suggest that adolescent and young adult (AYA) cancer survivors have a range of concerns regarding their fertility.
The female cancer survivors studied were more likely than males to describe feeling distressed and overwhelmed about their fertility.
Females also tended to worry about pregnancy-related health risks and cancer recurrence.
However, AYA cancer survivors of both sexes expressed concerns about genetic risk factors and how infertility might impact their future lives.
Catherine Benedict, PhD, of Memorial Sloan Kettering Cancer Center in New York, New York, and her colleagues conducted this research and reported the results in the Journal of Adolescent and Young Adult Oncology.
The researchers assessed fertility concerns in 43 AYA cancer survivors. They were 16 to 24 at the time of the study, had been diagnosed with cancer between ages 14 and 18, and were at least 6 months post-treatment.
The subjects either completed an individual interview (n=26) or participated in 1 of 4 focus groups (n=17).
Before treatment, 5 of the males had banked sperm, but none of the females took steps to preserve their fertility. More males (50%) than females (39%) reported uncertainty about their fertility.
The researchers identified 3 themes when discussing fertility with the study subjects: fertility concerns, emotions raised when discussing fertility, and strategies used to manage fertility concerns.
Concerns
Some subjects expressed concerns about how potential infertility could affect dating and relationships with partners (8% of males and 20% of females).
Some subjects were concerned about the health risks associated with having children—both risks to the subjects themselves and to any potential children (39% of males and 30% of females).
And some subjects were concerned about how potential infertility would affect their lives going forward (31% of males and 20% of females).
Emotions
When it came to emotions associated with fertility discussions, some subjects said they felt distressed and overwhelmed (23% of males and 30% of females).
However, some subjects said they weren’t concerned about their fertility (31% of males and 15% of females) or they felt hopeful despite the risk of infertility (8% of males and 20% of females).
Managing concerns
The subjects also mentioned a few strategies for managing fertility concerns. Some said they had accepted infertility (23% of males and 15% of females).
Some subjects said they weren’t going to worry about their fertility until they were older and actually wanted to have children (23% of males and 20% of females).
And some subjects said they would rely on assisted reproductive technology if necessary (31% of males and 20% of females).
Dr Benedict and her colleagues said this study suggests AYA cancer survivors may have a number of reproductive concerns and fertility-related distress, which may affect other areas of psychosocial functioning.
So future research should explore how to best incorporate fertility-related informational and support services more fully into survivorship care.

patient and her father
Photo by Rhoda Baer
Results of a small study suggest that adolescent and young adult (AYA) cancer survivors have a range of concerns regarding their fertility.
The female cancer survivors studied were more likely than males to describe feeling distressed and overwhelmed about their fertility.
Females also tended to worry about pregnancy-related health risks and cancer recurrence.
However, AYA cancer survivors of both sexes expressed concerns about genetic risk factors and how infertility might impact their future lives.
Catherine Benedict, PhD, of Memorial Sloan Kettering Cancer Center in New York, New York, and her colleagues conducted this research and reported the results in the Journal of Adolescent and Young Adult Oncology.
The researchers assessed fertility concerns in 43 AYA cancer survivors. They were 16 to 24 at the time of the study, had been diagnosed with cancer between ages 14 and 18, and were at least 6 months post-treatment.
The subjects either completed an individual interview (n=26) or participated in 1 of 4 focus groups (n=17).
Before treatment, 5 of the males had banked sperm, but none of the females took steps to preserve their fertility. More males (50%) than females (39%) reported uncertainty about their fertility.
The researchers identified 3 themes when discussing fertility with the study subjects: fertility concerns, emotions raised when discussing fertility, and strategies used to manage fertility concerns.
Concerns
Some subjects expressed concerns about how potential infertility could affect dating and relationships with partners (8% of males and 20% of females).
Some subjects were concerned about the health risks associated with having children—both risks to the subjects themselves and to any potential children (39% of males and 30% of females).
And some subjects were concerned about how potential infertility would affect their lives going forward (31% of males and 20% of females).
Emotions
When it came to emotions associated with fertility discussions, some subjects said they felt distressed and overwhelmed (23% of males and 30% of females).
However, some subjects said they weren’t concerned about their fertility (31% of males and 15% of females) or they felt hopeful despite the risk of infertility (8% of males and 20% of females).
Managing concerns
The subjects also mentioned a few strategies for managing fertility concerns. Some said they had accepted infertility (23% of males and 15% of females).
Some subjects said they weren’t going to worry about their fertility until they were older and actually wanted to have children (23% of males and 20% of females).
And some subjects said they would rely on assisted reproductive technology if necessary (31% of males and 20% of females).
Dr Benedict and her colleagues said this study suggests AYA cancer survivors may have a number of reproductive concerns and fertility-related distress, which may affect other areas of psychosocial functioning.
So future research should explore how to best incorporate fertility-related informational and support services more fully into survivorship care.
End-of-life cancer care by country
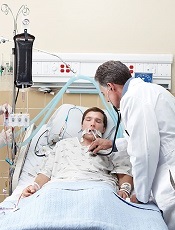
in the intensive care unit
A study of end-of-life cancer care practices in 7 countries suggests the US has the lowest proportion of deaths in the hospital and the lowest number of days in the hospital for patients in their last 6 months of life.
However, the US performed poorly in other aspects of care, particularly intensive care unit admissions and hospital expenditures.
The other countries included in the study were Belgium, Canada, England, Germany, the Netherlands, and Norway.
The research was published in JAMA.
Ezekiel J. Emanuel, MD, PhD, of the University of Pennsylvania in Philadelphia, and his colleagues examined patterns of care, healthcare utilization, and expenditures for dying cancer patients in the 7 aforementioned countries.
The researchers first analyzed data from 2010 that included subjects older than 65 years of age who died with cancer.
The proportion of patients who died in the hospital was 22.2% in the US, 29.4% in the Netherlands, 38.3% in Germany, 41.7% in England, 44.7% in Norway, 51.2% in Belgium, and 52.1% in Canada.
In the last 180 days of life, the mean number of days in the hospital per capita was 27.7 in Belgium, 24.8 in Norway, 21.7 in Germany, 19 in Canada, 18.3 in England, 17.8 in the Netherlands, and 10.7 in the US.
The proportion of patients admitted to the intensive care unit in their last 180 days of life was 40.3% in the US, 18.5% in Belgium, 15.2% in Canada, 10.2% in the Netherlands, and 8.2% in Germany. Data were not available for England and Norway.
In the last 180 days of life, average per capita hospital expenditures (in USD) were higher in Canada ($21,840), Norway ($19,783), and the US ($18,500), intermediate in Germany ($16,221) and Belgium ($15,699), and lowest in the Netherlands ($10,936) and England ($9342).
Analyses that included decedents of any age, decedents older than 65 years of age with lung cancer, and decedents older than 65 years in the US and Germany from 2012 showed similar results.
The researchers said this suggests the differences observed were driven more by end-of-life care practices and organization rather than differences in cohort identification.

in the intensive care unit
A study of end-of-life cancer care practices in 7 countries suggests the US has the lowest proportion of deaths in the hospital and the lowest number of days in the hospital for patients in their last 6 months of life.
However, the US performed poorly in other aspects of care, particularly intensive care unit admissions and hospital expenditures.
The other countries included in the study were Belgium, Canada, England, Germany, the Netherlands, and Norway.
The research was published in JAMA.
Ezekiel J. Emanuel, MD, PhD, of the University of Pennsylvania in Philadelphia, and his colleagues examined patterns of care, healthcare utilization, and expenditures for dying cancer patients in the 7 aforementioned countries.
The researchers first analyzed data from 2010 that included subjects older than 65 years of age who died with cancer.
The proportion of patients who died in the hospital was 22.2% in the US, 29.4% in the Netherlands, 38.3% in Germany, 41.7% in England, 44.7% in Norway, 51.2% in Belgium, and 52.1% in Canada.
In the last 180 days of life, the mean number of days in the hospital per capita was 27.7 in Belgium, 24.8 in Norway, 21.7 in Germany, 19 in Canada, 18.3 in England, 17.8 in the Netherlands, and 10.7 in the US.
The proportion of patients admitted to the intensive care unit in their last 180 days of life was 40.3% in the US, 18.5% in Belgium, 15.2% in Canada, 10.2% in the Netherlands, and 8.2% in Germany. Data were not available for England and Norway.
In the last 180 days of life, average per capita hospital expenditures (in USD) were higher in Canada ($21,840), Norway ($19,783), and the US ($18,500), intermediate in Germany ($16,221) and Belgium ($15,699), and lowest in the Netherlands ($10,936) and England ($9342).
Analyses that included decedents of any age, decedents older than 65 years of age with lung cancer, and decedents older than 65 years in the US and Germany from 2012 showed similar results.
The researchers said this suggests the differences observed were driven more by end-of-life care practices and organization rather than differences in cohort identification.

in the intensive care unit
A study of end-of-life cancer care practices in 7 countries suggests the US has the lowest proportion of deaths in the hospital and the lowest number of days in the hospital for patients in their last 6 months of life.
However, the US performed poorly in other aspects of care, particularly intensive care unit admissions and hospital expenditures.
The other countries included in the study were Belgium, Canada, England, Germany, the Netherlands, and Norway.
The research was published in JAMA.
Ezekiel J. Emanuel, MD, PhD, of the University of Pennsylvania in Philadelphia, and his colleagues examined patterns of care, healthcare utilization, and expenditures for dying cancer patients in the 7 aforementioned countries.
The researchers first analyzed data from 2010 that included subjects older than 65 years of age who died with cancer.
The proportion of patients who died in the hospital was 22.2% in the US, 29.4% in the Netherlands, 38.3% in Germany, 41.7% in England, 44.7% in Norway, 51.2% in Belgium, and 52.1% in Canada.
In the last 180 days of life, the mean number of days in the hospital per capita was 27.7 in Belgium, 24.8 in Norway, 21.7 in Germany, 19 in Canada, 18.3 in England, 17.8 in the Netherlands, and 10.7 in the US.
The proportion of patients admitted to the intensive care unit in their last 180 days of life was 40.3% in the US, 18.5% in Belgium, 15.2% in Canada, 10.2% in the Netherlands, and 8.2% in Germany. Data were not available for England and Norway.
In the last 180 days of life, average per capita hospital expenditures (in USD) were higher in Canada ($21,840), Norway ($19,783), and the US ($18,500), intermediate in Germany ($16,221) and Belgium ($15,699), and lowest in the Netherlands ($10,936) and England ($9342).
Analyses that included decedents of any age, decedents older than 65 years of age with lung cancer, and decedents older than 65 years in the US and Germany from 2012 showed similar results.
The researchers said this suggests the differences observed were driven more by end-of-life care practices and organization rather than differences in cohort identification.
Research helps explain how RBCs move

Scientists say they have determined how red blood cells (RBCs) move, showing that RBCs can be moved by external forces and actively “wriggle” on their own.
Linking physical principles and biological reality, the team found that fast molecules in the vicinity of RBCs make the cell membranes wriggle, but the cells themselves also become active when they have enough reaction time.
The group recounted these findings in Nature Physics.
Previously, scientists had only shown that RBCs’ constant wriggling was caused by external forces. But biological considerations suggested that internal forces might also be responsible for the RBCs’ membranes changing shape.
“So we started with the following question, ‘As blood cells are living cells, why shouldn’t internal forces inside the cell also have an impact on the membrane?’” said study author Timo Betz, PhD, of Münster University in Münster, Germany.
“For biologists, this is all clear, but these forces were just never a part of any physical equation.”
Dr Betz and his colleagues wanted to find out more about the mechanics of blood cells and gain a detailed understanding of the forces that move and shape cells.
The team said it is important to learn about RBCs’ properties and their internal forces because they are unusually soft and elastic and must change their shape to pass through blood vessels. It is precisely because RBCs are normally so soft that, in previous studies, physicists measured large thermal fluctuations at the outer membrane of the cells.
These natural movements of molecules are defined by the ambient temperature. In other words, the cell membrane moves because molecules in the vicinity jog it. Under the microscope, this makes the RBCs appear to be wriggling.
Although this explains why RBCs move, it does not address the question of possible internal forces being a contributing factor.
So Dr Betz and his colleagues used optical tweezers to take a close look at the fluctuations of RBCs. The team stretched RBCs in a petri dish and analyzed the behavior of the cells.
The result was that, if the RBCs had enough reaction time, they became active themselves and were able to counteract the force of the optical tweezers. If they did not have this time, they were at the mercy of their environment, and only temperature-related forces were measured.
“By comparing both sets of measurements, we can exactly define how fast the cells become active themselves and what force they generate in order to change shape,” Dr Betz explained.
He and his colleagues have a theory as to which forces inside RBCs cause the cell membrane to change shape.
“Transport proteins could generate such forces in the membrane by moving ions from one side of the membrane to the other,” said study author Gerhard Gompper, PhD, of the Jülich Institute of Complex Systems in Jülich, Germany.
“Now, it’s up to the biologists, because we physicists only have a rough idea about which proteins might be the drivers for this movement,” Dr Betz added. “On the other hand, we can predict exactly how fast and how strong they are.”

Scientists say they have determined how red blood cells (RBCs) move, showing that RBCs can be moved by external forces and actively “wriggle” on their own.
Linking physical principles and biological reality, the team found that fast molecules in the vicinity of RBCs make the cell membranes wriggle, but the cells themselves also become active when they have enough reaction time.
The group recounted these findings in Nature Physics.
Previously, scientists had only shown that RBCs’ constant wriggling was caused by external forces. But biological considerations suggested that internal forces might also be responsible for the RBCs’ membranes changing shape.
“So we started with the following question, ‘As blood cells are living cells, why shouldn’t internal forces inside the cell also have an impact on the membrane?’” said study author Timo Betz, PhD, of Münster University in Münster, Germany.
“For biologists, this is all clear, but these forces were just never a part of any physical equation.”
Dr Betz and his colleagues wanted to find out more about the mechanics of blood cells and gain a detailed understanding of the forces that move and shape cells.
The team said it is important to learn about RBCs’ properties and their internal forces because they are unusually soft and elastic and must change their shape to pass through blood vessels. It is precisely because RBCs are normally so soft that, in previous studies, physicists measured large thermal fluctuations at the outer membrane of the cells.
These natural movements of molecules are defined by the ambient temperature. In other words, the cell membrane moves because molecules in the vicinity jog it. Under the microscope, this makes the RBCs appear to be wriggling.
Although this explains why RBCs move, it does not address the question of possible internal forces being a contributing factor.
So Dr Betz and his colleagues used optical tweezers to take a close look at the fluctuations of RBCs. The team stretched RBCs in a petri dish and analyzed the behavior of the cells.
The result was that, if the RBCs had enough reaction time, they became active themselves and were able to counteract the force of the optical tweezers. If they did not have this time, they were at the mercy of their environment, and only temperature-related forces were measured.
“By comparing both sets of measurements, we can exactly define how fast the cells become active themselves and what force they generate in order to change shape,” Dr Betz explained.
He and his colleagues have a theory as to which forces inside RBCs cause the cell membrane to change shape.
“Transport proteins could generate such forces in the membrane by moving ions from one side of the membrane to the other,” said study author Gerhard Gompper, PhD, of the Jülich Institute of Complex Systems in Jülich, Germany.
“Now, it’s up to the biologists, because we physicists only have a rough idea about which proteins might be the drivers for this movement,” Dr Betz added. “On the other hand, we can predict exactly how fast and how strong they are.”

Scientists say they have determined how red blood cells (RBCs) move, showing that RBCs can be moved by external forces and actively “wriggle” on their own.
Linking physical principles and biological reality, the team found that fast molecules in the vicinity of RBCs make the cell membranes wriggle, but the cells themselves also become active when they have enough reaction time.
The group recounted these findings in Nature Physics.
Previously, scientists had only shown that RBCs’ constant wriggling was caused by external forces. But biological considerations suggested that internal forces might also be responsible for the RBCs’ membranes changing shape.
“So we started with the following question, ‘As blood cells are living cells, why shouldn’t internal forces inside the cell also have an impact on the membrane?’” said study author Timo Betz, PhD, of Münster University in Münster, Germany.
“For biologists, this is all clear, but these forces were just never a part of any physical equation.”
Dr Betz and his colleagues wanted to find out more about the mechanics of blood cells and gain a detailed understanding of the forces that move and shape cells.
The team said it is important to learn about RBCs’ properties and their internal forces because they are unusually soft and elastic and must change their shape to pass through blood vessels. It is precisely because RBCs are normally so soft that, in previous studies, physicists measured large thermal fluctuations at the outer membrane of the cells.
These natural movements of molecules are defined by the ambient temperature. In other words, the cell membrane moves because molecules in the vicinity jog it. Under the microscope, this makes the RBCs appear to be wriggling.
Although this explains why RBCs move, it does not address the question of possible internal forces being a contributing factor.
So Dr Betz and his colleagues used optical tweezers to take a close look at the fluctuations of RBCs. The team stretched RBCs in a petri dish and analyzed the behavior of the cells.
The result was that, if the RBCs had enough reaction time, they became active themselves and were able to counteract the force of the optical tweezers. If they did not have this time, they were at the mercy of their environment, and only temperature-related forces were measured.
“By comparing both sets of measurements, we can exactly define how fast the cells become active themselves and what force they generate in order to change shape,” Dr Betz explained.
He and his colleagues have a theory as to which forces inside RBCs cause the cell membrane to change shape.
“Transport proteins could generate such forces in the membrane by moving ions from one side of the membrane to the other,” said study author Gerhard Gompper, PhD, of the Jülich Institute of Complex Systems in Jülich, Germany.
“Now, it’s up to the biologists, because we physicists only have a rough idea about which proteins might be the drivers for this movement,” Dr Betz added. “On the other hand, we can predict exactly how fast and how strong they are.”