User login
Group ranks TKIs according to cardiotoxicity
Researchers say they have devised a way to rank tyrosine kinase inhibitors (TKIs) based on their likelihood of causing lasting heart damage in patients.
The team found they could accurately identify those TKIs already known to be the most dangerous.
The researchers believe that, in the future, their system may prove useful in the early stages of drug development to screen new compounds for cardiotoxicity.
“This type of study represents a critical step forward from the usual process running from initial drug discovery and clinical trials in human patients,” said study author Joseph Wu, MD, PhD, of Stanford University School of Medicine in California.
“It will help pharmaceutical companies better focus their efforts on developing safer drugs, and it will provide patients more effective drugs with fewer side effects.”
Dr Wu and his colleagues described this research in Science Translational Medicine.
The researchers began with human induced pluripotent stem cell-derived cardiomyocytes generated from the cells of 11 healthy people and 2 patients with kidney cancer.
The team grew the cardiomyocytes in a dish and tested the effects of 21 commonly used TKIs on the cells.
Treatment with drug levels equivalent to those taken by patients often caused the cells to beat irregularly and begin to die. The cells also displayed differences in the electrophysiological signaling that controls their contraction.
The researchers used these and other measurements to develop a cardiac safety index for each drug. They found that TKIs known to be particularly dangerous to heart function had the lowest safety indices, and TKIs known to be better tolerated by patients ranked higher on the safety index.
High cardiotoxicity
Seven of the 21 TKIs tested were assigned cardiac safety indices at or below 0.1—the threshold limit at which the researchers designated a drug highly cardiotoxic.
These TKIs (from lowest to highest safety score) were vemurafenib, sorafenib, doxorubicin, regorafenib, vandetanib, crizotinib, and nilotinib.
Three of the most cardiotoxic TKIs (regorafenib, sorafenib, and vandetanib) are known to inhibit the same 2 signaling pathways: VEGFR2 and PDGFR.
The researchers noticed that cells treated with these 3 drugs ramped up the activity of a cellular signaling pathway that responds to insulin or IGF1, an insulin-like growth factor.
This discovery, coupled with the fact that treatment with insulin or IGF1 is known to enhance heart function during adverse cardiac events such as heart attacks, led the researchers to experiment further.
They found that exposing the cells to insulin or IGF1 made it less likely they would die due to TKIs blocking the VEGFR2 and PDGFR pathways. Although more research is needed, these findings suggest it may be possible to alleviate some of the heart damage in patients receiving these TKIs.
“There is a critical need for a way to ‘safety test’ all drugs earlier in development before they are administered to patients,” Dr Wu said. “Our drug safety index is a step in that direction.” ![]()
Researchers say they have devised a way to rank tyrosine kinase inhibitors (TKIs) based on their likelihood of causing lasting heart damage in patients.
The team found they could accurately identify those TKIs already known to be the most dangerous.
The researchers believe that, in the future, their system may prove useful in the early stages of drug development to screen new compounds for cardiotoxicity.
“This type of study represents a critical step forward from the usual process running from initial drug discovery and clinical trials in human patients,” said study author Joseph Wu, MD, PhD, of Stanford University School of Medicine in California.
“It will help pharmaceutical companies better focus their efforts on developing safer drugs, and it will provide patients more effective drugs with fewer side effects.”
Dr Wu and his colleagues described this research in Science Translational Medicine.
The researchers began with human induced pluripotent stem cell-derived cardiomyocytes generated from the cells of 11 healthy people and 2 patients with kidney cancer.
The team grew the cardiomyocytes in a dish and tested the effects of 21 commonly used TKIs on the cells.
Treatment with drug levels equivalent to those taken by patients often caused the cells to beat irregularly and begin to die. The cells also displayed differences in the electrophysiological signaling that controls their contraction.
The researchers used these and other measurements to develop a cardiac safety index for each drug. They found that TKIs known to be particularly dangerous to heart function had the lowest safety indices, and TKIs known to be better tolerated by patients ranked higher on the safety index.
High cardiotoxicity
Seven of the 21 TKIs tested were assigned cardiac safety indices at or below 0.1—the threshold limit at which the researchers designated a drug highly cardiotoxic.
These TKIs (from lowest to highest safety score) were vemurafenib, sorafenib, doxorubicin, regorafenib, vandetanib, crizotinib, and nilotinib.
Three of the most cardiotoxic TKIs (regorafenib, sorafenib, and vandetanib) are known to inhibit the same 2 signaling pathways: VEGFR2 and PDGFR.
The researchers noticed that cells treated with these 3 drugs ramped up the activity of a cellular signaling pathway that responds to insulin or IGF1, an insulin-like growth factor.
This discovery, coupled with the fact that treatment with insulin or IGF1 is known to enhance heart function during adverse cardiac events such as heart attacks, led the researchers to experiment further.
They found that exposing the cells to insulin or IGF1 made it less likely they would die due to TKIs blocking the VEGFR2 and PDGFR pathways. Although more research is needed, these findings suggest it may be possible to alleviate some of the heart damage in patients receiving these TKIs.
“There is a critical need for a way to ‘safety test’ all drugs earlier in development before they are administered to patients,” Dr Wu said. “Our drug safety index is a step in that direction.” ![]()
Researchers say they have devised a way to rank tyrosine kinase inhibitors (TKIs) based on their likelihood of causing lasting heart damage in patients.
The team found they could accurately identify those TKIs already known to be the most dangerous.
The researchers believe that, in the future, their system may prove useful in the early stages of drug development to screen new compounds for cardiotoxicity.
“This type of study represents a critical step forward from the usual process running from initial drug discovery and clinical trials in human patients,” said study author Joseph Wu, MD, PhD, of Stanford University School of Medicine in California.
“It will help pharmaceutical companies better focus their efforts on developing safer drugs, and it will provide patients more effective drugs with fewer side effects.”
Dr Wu and his colleagues described this research in Science Translational Medicine.
The researchers began with human induced pluripotent stem cell-derived cardiomyocytes generated from the cells of 11 healthy people and 2 patients with kidney cancer.
The team grew the cardiomyocytes in a dish and tested the effects of 21 commonly used TKIs on the cells.
Treatment with drug levels equivalent to those taken by patients often caused the cells to beat irregularly and begin to die. The cells also displayed differences in the electrophysiological signaling that controls their contraction.
The researchers used these and other measurements to develop a cardiac safety index for each drug. They found that TKIs known to be particularly dangerous to heart function had the lowest safety indices, and TKIs known to be better tolerated by patients ranked higher on the safety index.
High cardiotoxicity
Seven of the 21 TKIs tested were assigned cardiac safety indices at or below 0.1—the threshold limit at which the researchers designated a drug highly cardiotoxic.
These TKIs (from lowest to highest safety score) were vemurafenib, sorafenib, doxorubicin, regorafenib, vandetanib, crizotinib, and nilotinib.
Three of the most cardiotoxic TKIs (regorafenib, sorafenib, and vandetanib) are known to inhibit the same 2 signaling pathways: VEGFR2 and PDGFR.
The researchers noticed that cells treated with these 3 drugs ramped up the activity of a cellular signaling pathway that responds to insulin or IGF1, an insulin-like growth factor.
This discovery, coupled with the fact that treatment with insulin or IGF1 is known to enhance heart function during adverse cardiac events such as heart attacks, led the researchers to experiment further.
They found that exposing the cells to insulin or IGF1 made it less likely they would die due to TKIs blocking the VEGFR2 and PDGFR pathways. Although more research is needed, these findings suggest it may be possible to alleviate some of the heart damage in patients receiving these TKIs.
“There is a critical need for a way to ‘safety test’ all drugs earlier in development before they are administered to patients,” Dr Wu said. “Our drug safety index is a step in that direction.” ![]()
Magnetic implant may offer new drug delivery method
A tiny magnetic implant could provide a new method of drug delivery, according to research published in Advanced Functional Materials.
The device is a silicone sponge with magnetic carbonyl iron particles wrapped in a round polymer layer. It measures 6 mm in diameter.
A drug is injected into the device, which is surgically implanted in the area being treated.
Passing a magnet over the implant activates the device by deforming the sponge and triggering the release of the drug into surrounding tissue through a tiny opening.
“Drug implants can be safe and effective for treating many conditions, and magnetically controlled implants are particularly interesting because you can adjust the dose after implantation by using different magnet strengths,” said study author Ali Shademani, a PhD student at the University of British Columbia (UBC) in Vancouver, British Columbia, Canada.
“This device lets you release the actual dose that the patient needs when they need it, and it’s sufficiently easy to use that patients could administer their own medication one day without having to go to a hospital,” added John K. Jackson, also of UBC.
The researchers tested the device on animal tissue in the lab using the prostate cancer drug docetaxel. The device was able to deliver the drug on demand even after repeated use.
The drug also produced an effect on cancer cells comparable to that of freshly administered docetaxel, suggesting that drugs stored in the device stay effective.
The researchers are now working on refining the device and narrowing down the conditions for its use.
“This could one day be used for administering painkillers, hormones, chemotherapy drugs, and other treatments for a wide range of health conditions,” said Mu Chiao, PhD, of UBC. “In the next few years, we hope to be able to test it for long-term use and for viability in living models.” ![]()
A tiny magnetic implant could provide a new method of drug delivery, according to research published in Advanced Functional Materials.
The device is a silicone sponge with magnetic carbonyl iron particles wrapped in a round polymer layer. It measures 6 mm in diameter.
A drug is injected into the device, which is surgically implanted in the area being treated.
Passing a magnet over the implant activates the device by deforming the sponge and triggering the release of the drug into surrounding tissue through a tiny opening.
“Drug implants can be safe and effective for treating many conditions, and magnetically controlled implants are particularly interesting because you can adjust the dose after implantation by using different magnet strengths,” said study author Ali Shademani, a PhD student at the University of British Columbia (UBC) in Vancouver, British Columbia, Canada.
“This device lets you release the actual dose that the patient needs when they need it, and it’s sufficiently easy to use that patients could administer their own medication one day without having to go to a hospital,” added John K. Jackson, also of UBC.
The researchers tested the device on animal tissue in the lab using the prostate cancer drug docetaxel. The device was able to deliver the drug on demand even after repeated use.
The drug also produced an effect on cancer cells comparable to that of freshly administered docetaxel, suggesting that drugs stored in the device stay effective.
The researchers are now working on refining the device and narrowing down the conditions for its use.
“This could one day be used for administering painkillers, hormones, chemotherapy drugs, and other treatments for a wide range of health conditions,” said Mu Chiao, PhD, of UBC. “In the next few years, we hope to be able to test it for long-term use and for viability in living models.” ![]()
A tiny magnetic implant could provide a new method of drug delivery, according to research published in Advanced Functional Materials.
The device is a silicone sponge with magnetic carbonyl iron particles wrapped in a round polymer layer. It measures 6 mm in diameter.
A drug is injected into the device, which is surgically implanted in the area being treated.
Passing a magnet over the implant activates the device by deforming the sponge and triggering the release of the drug into surrounding tissue through a tiny opening.
“Drug implants can be safe and effective for treating many conditions, and magnetically controlled implants are particularly interesting because you can adjust the dose after implantation by using different magnet strengths,” said study author Ali Shademani, a PhD student at the University of British Columbia (UBC) in Vancouver, British Columbia, Canada.
“This device lets you release the actual dose that the patient needs when they need it, and it’s sufficiently easy to use that patients could administer their own medication one day without having to go to a hospital,” added John K. Jackson, also of UBC.
The researchers tested the device on animal tissue in the lab using the prostate cancer drug docetaxel. The device was able to deliver the drug on demand even after repeated use.
The drug also produced an effect on cancer cells comparable to that of freshly administered docetaxel, suggesting that drugs stored in the device stay effective.
The researchers are now working on refining the device and narrowing down the conditions for its use.
“This could one day be used for administering painkillers, hormones, chemotherapy drugs, and other treatments for a wide range of health conditions,” said Mu Chiao, PhD, of UBC. “In the next few years, we hope to be able to test it for long-term use and for viability in living models.” ![]()
Vaccination approach prevents malaria
A novel vaccination approach can offer 100% protection against malaria, according to research published in Nature.
The approach involved direct venous inoculation of aseptic, purified, cryopreserved, non-irradiated Plasmodium falciparum sporozoites (PfSPZ) in healthy adults taking chloroquine for antimalarial chemoprophylaxis (PfSPZ-CVac).
At the optimal dose and schedule, PfSPZ-CVac prevented infection in 9 of 9 volunteers, and this protection lasted 10 weeks after the last dose.
In addition, researchers said PfSPZ-CVac was well-tolerated.
PfSPZ was provided by Sanaria. This research was supported by federal funding and the Bill and Melinda Gates Foundation.
Treatment and challenge
The researchers tested PfSPZ-CVac in healthy adults, ages 18 to 45, who were malaria-naive. They received different doses of PfSPZ at different intervals and different doses/schedules of chloroquine.
In the first part of the study, test subjects received 3 doses of PfSPZ at 3.2 × 103 (n=9), 1.28 × 104 (n=9), or 5.12 × 104 (n=9), and controls (n=13) received 3 doses of normal saline.
Doses were given at 28-day intervals, and controlled human malaria infection (CHMI) was performed at 8 to 10 weeks after immunization.
In the second part of the study, test subjects received 3 doses of PfSPZ at 5.12 × 104 at 14-day intervals (n=9) or 5-day intervals (n=9), and controls (n=6) received 3 doses of normal saline.
CHMI was performed at 10 weeks post-immunization.
Most subjects received chloroquine at a base loading dose of 10 mg kg-1 or 620 mg two days before the first dose of PfSPZ, whichever dose was less, followed by weekly doses of 5 mg kg-1 or 310 mg through 5 days after the last dose of PfSPZ. Subjects who were immunized on days 0, 5, and 10 received chloroquine on days 0, 5, 10, and 15.
Results
All controls, who received normal saline plus chloroquine, developed parasitemia.
However, 3 doses of PfSPZ at 5.12 × 104, given at 28-day intervals, prevented infection in 9 of 9 subjects (100%).
The 3.2 × 103 dose of PfSPZ (also given at 28-day intervals) protected 3 of 9 subjects (33%), and the 1.28 × 104 dose protected 6 of 9 subjects (67%).
Three doses of PfSPZ at 5.12 × 104, given at 5-day intervals, protected 5 of 8 subjects (63%). And 3 doses of PfSPZ at 5.12 × 104, given at 14-day intervals, protected 6 of 9 subjects (67%).
“By vaccinating with a live, fully active pathogen, it seems clear that we were able to set off a very strong immune response,” said study author Benjamin Mordmüller, of the University of Tübingen in Germany.
“Additionally, all the data we have so far indicate that what we have here is relatively stable, long-lasting protection.”
“That protection was probably caused by specific T lymphocytes and antibody responses to the parasites in the liver,” added Peter Kremsner, also of the University of Tübingen.
The researchers said there were no serious adverse events, and the frequencies of grade 1-3 events were similar in subjects who received PfSPZ-CVac and controls.
Chloroquine was considered well-tolerated, although 2 subjects discontinued the trial after the loading dose because they experienced nausea and vomiting. ![]()
A novel vaccination approach can offer 100% protection against malaria, according to research published in Nature.
The approach involved direct venous inoculation of aseptic, purified, cryopreserved, non-irradiated Plasmodium falciparum sporozoites (PfSPZ) in healthy adults taking chloroquine for antimalarial chemoprophylaxis (PfSPZ-CVac).
At the optimal dose and schedule, PfSPZ-CVac prevented infection in 9 of 9 volunteers, and this protection lasted 10 weeks after the last dose.
In addition, researchers said PfSPZ-CVac was well-tolerated.
PfSPZ was provided by Sanaria. This research was supported by federal funding and the Bill and Melinda Gates Foundation.
Treatment and challenge
The researchers tested PfSPZ-CVac in healthy adults, ages 18 to 45, who were malaria-naive. They received different doses of PfSPZ at different intervals and different doses/schedules of chloroquine.
In the first part of the study, test subjects received 3 doses of PfSPZ at 3.2 × 103 (n=9), 1.28 × 104 (n=9), or 5.12 × 104 (n=9), and controls (n=13) received 3 doses of normal saline.
Doses were given at 28-day intervals, and controlled human malaria infection (CHMI) was performed at 8 to 10 weeks after immunization.
In the second part of the study, test subjects received 3 doses of PfSPZ at 5.12 × 104 at 14-day intervals (n=9) or 5-day intervals (n=9), and controls (n=6) received 3 doses of normal saline.
CHMI was performed at 10 weeks post-immunization.
Most subjects received chloroquine at a base loading dose of 10 mg kg-1 or 620 mg two days before the first dose of PfSPZ, whichever dose was less, followed by weekly doses of 5 mg kg-1 or 310 mg through 5 days after the last dose of PfSPZ. Subjects who were immunized on days 0, 5, and 10 received chloroquine on days 0, 5, 10, and 15.
Results
All controls, who received normal saline plus chloroquine, developed parasitemia.
However, 3 doses of PfSPZ at 5.12 × 104, given at 28-day intervals, prevented infection in 9 of 9 subjects (100%).
The 3.2 × 103 dose of PfSPZ (also given at 28-day intervals) protected 3 of 9 subjects (33%), and the 1.28 × 104 dose protected 6 of 9 subjects (67%).
Three doses of PfSPZ at 5.12 × 104, given at 5-day intervals, protected 5 of 8 subjects (63%). And 3 doses of PfSPZ at 5.12 × 104, given at 14-day intervals, protected 6 of 9 subjects (67%).
“By vaccinating with a live, fully active pathogen, it seems clear that we were able to set off a very strong immune response,” said study author Benjamin Mordmüller, of the University of Tübingen in Germany.
“Additionally, all the data we have so far indicate that what we have here is relatively stable, long-lasting protection.”
“That protection was probably caused by specific T lymphocytes and antibody responses to the parasites in the liver,” added Peter Kremsner, also of the University of Tübingen.
The researchers said there were no serious adverse events, and the frequencies of grade 1-3 events were similar in subjects who received PfSPZ-CVac and controls.
Chloroquine was considered well-tolerated, although 2 subjects discontinued the trial after the loading dose because they experienced nausea and vomiting. ![]()
A novel vaccination approach can offer 100% protection against malaria, according to research published in Nature.
The approach involved direct venous inoculation of aseptic, purified, cryopreserved, non-irradiated Plasmodium falciparum sporozoites (PfSPZ) in healthy adults taking chloroquine for antimalarial chemoprophylaxis (PfSPZ-CVac).
At the optimal dose and schedule, PfSPZ-CVac prevented infection in 9 of 9 volunteers, and this protection lasted 10 weeks after the last dose.
In addition, researchers said PfSPZ-CVac was well-tolerated.
PfSPZ was provided by Sanaria. This research was supported by federal funding and the Bill and Melinda Gates Foundation.
Treatment and challenge
The researchers tested PfSPZ-CVac in healthy adults, ages 18 to 45, who were malaria-naive. They received different doses of PfSPZ at different intervals and different doses/schedules of chloroquine.
In the first part of the study, test subjects received 3 doses of PfSPZ at 3.2 × 103 (n=9), 1.28 × 104 (n=9), or 5.12 × 104 (n=9), and controls (n=13) received 3 doses of normal saline.
Doses were given at 28-day intervals, and controlled human malaria infection (CHMI) was performed at 8 to 10 weeks after immunization.
In the second part of the study, test subjects received 3 doses of PfSPZ at 5.12 × 104 at 14-day intervals (n=9) or 5-day intervals (n=9), and controls (n=6) received 3 doses of normal saline.
CHMI was performed at 10 weeks post-immunization.
Most subjects received chloroquine at a base loading dose of 10 mg kg-1 or 620 mg two days before the first dose of PfSPZ, whichever dose was less, followed by weekly doses of 5 mg kg-1 or 310 mg through 5 days after the last dose of PfSPZ. Subjects who were immunized on days 0, 5, and 10 received chloroquine on days 0, 5, 10, and 15.
Results
All controls, who received normal saline plus chloroquine, developed parasitemia.
However, 3 doses of PfSPZ at 5.12 × 104, given at 28-day intervals, prevented infection in 9 of 9 subjects (100%).
The 3.2 × 103 dose of PfSPZ (also given at 28-day intervals) protected 3 of 9 subjects (33%), and the 1.28 × 104 dose protected 6 of 9 subjects (67%).
Three doses of PfSPZ at 5.12 × 104, given at 5-day intervals, protected 5 of 8 subjects (63%). And 3 doses of PfSPZ at 5.12 × 104, given at 14-day intervals, protected 6 of 9 subjects (67%).
“By vaccinating with a live, fully active pathogen, it seems clear that we were able to set off a very strong immune response,” said study author Benjamin Mordmüller, of the University of Tübingen in Germany.
“Additionally, all the data we have so far indicate that what we have here is relatively stable, long-lasting protection.”
“That protection was probably caused by specific T lymphocytes and antibody responses to the parasites in the liver,” added Peter Kremsner, also of the University of Tübingen.
The researchers said there were no serious adverse events, and the frequencies of grade 1-3 events were similar in subjects who received PfSPZ-CVac and controls.
Chloroquine was considered well-tolerated, although 2 subjects discontinued the trial after the loading dose because they experienced nausea and vomiting. ![]()
Cancer survivors report pros and cons of telehealth

Photo by Daniel Sone
Cancer survivors report a range of benefits and detriments related to telehealth, according to research published in the Journal of Medical Internet Research.
Telehealth is the use of technology to provide remote, personalized healthcare to patients.
Telehealth services allow patients to have meetings and follow-up consultations with healthcare professionals either on the phone or through online services at a time that suits the patients.
Anna Cox, PhD, of the University of Surrey in the UK, and her colleagues examined 22 studies, published between 2006 and 2016, that reported cancer patients’ direct views on their experience of telehealth.
Some of the cancer survivors studied reported their appreciation of the flexibility and convenience of telehealth, which enabled them to engage with healthcare providers with minimum disruption to their lives and in a comfortable, familiar environment.
“Our research found that cancer survivors wanted to get back to their daily lives as quickly as possible,” Dr Cox said. “Telehealth helped facilitate this, as it removed the often burdensome visits to hospital and enabled the integration of care into daily routines.”
However, not all subjects viewed telehealth as a convenience. Of the Internet-based interventions studied, 2 were perceived as an extra burden, and 1 was considered too time-consuming.
In addition, some study participants viewed telehealth as an impersonal service that did not allow them to meet their healthcare team in person.
On the other hand, the invisibility and perceived anonymity that telehealth provided sometimes reduced cancer survivors’ sense of vulnerability and enabled them to raise concerns remotely that they would not have wanted to discuss face-to-face.
And, in 8 different studies, subjects said telehealth had educated them about ways they could improve or manage their symptoms, or it had raised their awareness of potential issues they might experience.
Unfortunately, some of the cancer survivors studied said they were unable to use telehealth due to personal circumstances, such as hearing issues and lack of computer literacy skills.
“For many cancer survivors, telehealth supported their independence and offered them reassurance,” Dr Cox noted. “However, it is all down to personal preference, as some cancer survivors still preferred traditional methods of care.” ![]()

Photo by Daniel Sone
Cancer survivors report a range of benefits and detriments related to telehealth, according to research published in the Journal of Medical Internet Research.
Telehealth is the use of technology to provide remote, personalized healthcare to patients.
Telehealth services allow patients to have meetings and follow-up consultations with healthcare professionals either on the phone or through online services at a time that suits the patients.
Anna Cox, PhD, of the University of Surrey in the UK, and her colleagues examined 22 studies, published between 2006 and 2016, that reported cancer patients’ direct views on their experience of telehealth.
Some of the cancer survivors studied reported their appreciation of the flexibility and convenience of telehealth, which enabled them to engage with healthcare providers with minimum disruption to their lives and in a comfortable, familiar environment.
“Our research found that cancer survivors wanted to get back to their daily lives as quickly as possible,” Dr Cox said. “Telehealth helped facilitate this, as it removed the often burdensome visits to hospital and enabled the integration of care into daily routines.”
However, not all subjects viewed telehealth as a convenience. Of the Internet-based interventions studied, 2 were perceived as an extra burden, and 1 was considered too time-consuming.
In addition, some study participants viewed telehealth as an impersonal service that did not allow them to meet their healthcare team in person.
On the other hand, the invisibility and perceived anonymity that telehealth provided sometimes reduced cancer survivors’ sense of vulnerability and enabled them to raise concerns remotely that they would not have wanted to discuss face-to-face.
And, in 8 different studies, subjects said telehealth had educated them about ways they could improve or manage their symptoms, or it had raised their awareness of potential issues they might experience.
Unfortunately, some of the cancer survivors studied said they were unable to use telehealth due to personal circumstances, such as hearing issues and lack of computer literacy skills.
“For many cancer survivors, telehealth supported their independence and offered them reassurance,” Dr Cox noted. “However, it is all down to personal preference, as some cancer survivors still preferred traditional methods of care.” ![]()

Photo by Daniel Sone
Cancer survivors report a range of benefits and detriments related to telehealth, according to research published in the Journal of Medical Internet Research.
Telehealth is the use of technology to provide remote, personalized healthcare to patients.
Telehealth services allow patients to have meetings and follow-up consultations with healthcare professionals either on the phone or through online services at a time that suits the patients.
Anna Cox, PhD, of the University of Surrey in the UK, and her colleagues examined 22 studies, published between 2006 and 2016, that reported cancer patients’ direct views on their experience of telehealth.
Some of the cancer survivors studied reported their appreciation of the flexibility and convenience of telehealth, which enabled them to engage with healthcare providers with minimum disruption to their lives and in a comfortable, familiar environment.
“Our research found that cancer survivors wanted to get back to their daily lives as quickly as possible,” Dr Cox said. “Telehealth helped facilitate this, as it removed the often burdensome visits to hospital and enabled the integration of care into daily routines.”
However, not all subjects viewed telehealth as a convenience. Of the Internet-based interventions studied, 2 were perceived as an extra burden, and 1 was considered too time-consuming.
In addition, some study participants viewed telehealth as an impersonal service that did not allow them to meet their healthcare team in person.
On the other hand, the invisibility and perceived anonymity that telehealth provided sometimes reduced cancer survivors’ sense of vulnerability and enabled them to raise concerns remotely that they would not have wanted to discuss face-to-face.
And, in 8 different studies, subjects said telehealth had educated them about ways they could improve or manage their symptoms, or it had raised their awareness of potential issues they might experience.
Unfortunately, some of the cancer survivors studied said they were unable to use telehealth due to personal circumstances, such as hearing issues and lack of computer literacy skills.
“For many cancer survivors, telehealth supported their independence and offered them reassurance,” Dr Cox noted. “However, it is all down to personal preference, as some cancer survivors still preferred traditional methods of care.” ![]()
Predicting the efficacy of malaria vaccines
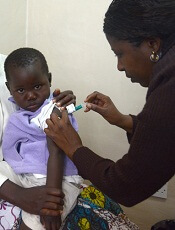
Photo by Caitlin Kleiboer
Researchers say they have identified molecular signatures that could potentially be used to predict whether the malaria vaccine RTS,S will be effective.
The group says the research, published in PNAS, could inform decisions on how RTS,S or other malaria vaccines are deployed or modified.
RTS,S (also known as RTS,S/AS01 or Mosquirix) has been shown to provide partial protection against malaria in phase 2 and phase 3 trials.
The vaccine is scheduled for roll-out through pilot projects in 3 African countries next year, according to the World Health Organization.
With the current study, researchers used a systems biology approach to identify molecular signatures induced after subjects were vaccinated with RTS,S.
The team looked at 2 groups of subjects:
- Individuals vaccinated with the standard RTS,S vaccination regimen, which consists of 3 RTS,S immunizations (RRR)
- Individuals vaccinated first with recombinant adenovirus 35 (Ad35) expressing the circumsporozoite malaria antigen, followed by 2 immunizations with RTS,S (ARR).
Vaccinated subjects were exposed to mosquitoes infected with Plasmodium falciparum 3 weeks after their final immunization.
Both vaccination regimens resulted in about 50% protection from malaria infection. And the researchers identified markers in each group that were associated with protection.
In the RRR group, circumsporozoite protein-specific antibody titers, prior to the challenge with infected mosquitoes, were associated with protection from malaria infection.
In addition, molecular signatures of B and plasma cells detected in peripheral blood mononuclear cells were associated with pre-challenge antibody titers and protection from malaria infection in the RRR group.
In the ARR group, protection from infection was associated with polyfunctional CD4+ T-cell responses 2 weeks after priming with Ad35, early signatures of innate immunity, and dendritic cell activation.
In both groups, natural killer (NK) cell signatures were negatively correlated with protection.
“Many of the genes contained in the predictive signatures are known to be expressed in natural killer cells, which mediate critical immune functions against viruses,” said study author Bali Pulendran, PhD, of Emory University School of Medicine in Atlanta, Georgia.
“It was a surprise to see such a robust ‘NK cell signature’ in predicting success of vaccination against the malaria parasite and raises the hypothesis that such cells may be playing a vital role in orchestrating immunity against malaria.”
Dr Pulendran and his colleagues said the results of this research suggest protective immunity against P falciparum can be achieved via multiple mechanisms.
“The extent to which these candidate signatures of protection can successfully predict vaccine efficacy in other field trials remain to be determined,” Dr Pulendran noted. ![]()

Photo by Caitlin Kleiboer
Researchers say they have identified molecular signatures that could potentially be used to predict whether the malaria vaccine RTS,S will be effective.
The group says the research, published in PNAS, could inform decisions on how RTS,S or other malaria vaccines are deployed or modified.
RTS,S (also known as RTS,S/AS01 or Mosquirix) has been shown to provide partial protection against malaria in phase 2 and phase 3 trials.
The vaccine is scheduled for roll-out through pilot projects in 3 African countries next year, according to the World Health Organization.
With the current study, researchers used a systems biology approach to identify molecular signatures induced after subjects were vaccinated with RTS,S.
The team looked at 2 groups of subjects:
- Individuals vaccinated with the standard RTS,S vaccination regimen, which consists of 3 RTS,S immunizations (RRR)
- Individuals vaccinated first with recombinant adenovirus 35 (Ad35) expressing the circumsporozoite malaria antigen, followed by 2 immunizations with RTS,S (ARR).
Vaccinated subjects were exposed to mosquitoes infected with Plasmodium falciparum 3 weeks after their final immunization.
Both vaccination regimens resulted in about 50% protection from malaria infection. And the researchers identified markers in each group that were associated with protection.
In the RRR group, circumsporozoite protein-specific antibody titers, prior to the challenge with infected mosquitoes, were associated with protection from malaria infection.
In addition, molecular signatures of B and plasma cells detected in peripheral blood mononuclear cells were associated with pre-challenge antibody titers and protection from malaria infection in the RRR group.
In the ARR group, protection from infection was associated with polyfunctional CD4+ T-cell responses 2 weeks after priming with Ad35, early signatures of innate immunity, and dendritic cell activation.
In both groups, natural killer (NK) cell signatures were negatively correlated with protection.
“Many of the genes contained in the predictive signatures are known to be expressed in natural killer cells, which mediate critical immune functions against viruses,” said study author Bali Pulendran, PhD, of Emory University School of Medicine in Atlanta, Georgia.
“It was a surprise to see such a robust ‘NK cell signature’ in predicting success of vaccination against the malaria parasite and raises the hypothesis that such cells may be playing a vital role in orchestrating immunity against malaria.”
Dr Pulendran and his colleagues said the results of this research suggest protective immunity against P falciparum can be achieved via multiple mechanisms.
“The extent to which these candidate signatures of protection can successfully predict vaccine efficacy in other field trials remain to be determined,” Dr Pulendran noted. ![]()

Photo by Caitlin Kleiboer
Researchers say they have identified molecular signatures that could potentially be used to predict whether the malaria vaccine RTS,S will be effective.
The group says the research, published in PNAS, could inform decisions on how RTS,S or other malaria vaccines are deployed or modified.
RTS,S (also known as RTS,S/AS01 or Mosquirix) has been shown to provide partial protection against malaria in phase 2 and phase 3 trials.
The vaccine is scheduled for roll-out through pilot projects in 3 African countries next year, according to the World Health Organization.
With the current study, researchers used a systems biology approach to identify molecular signatures induced after subjects were vaccinated with RTS,S.
The team looked at 2 groups of subjects:
- Individuals vaccinated with the standard RTS,S vaccination regimen, which consists of 3 RTS,S immunizations (RRR)
- Individuals vaccinated first with recombinant adenovirus 35 (Ad35) expressing the circumsporozoite malaria antigen, followed by 2 immunizations with RTS,S (ARR).
Vaccinated subjects were exposed to mosquitoes infected with Plasmodium falciparum 3 weeks after their final immunization.
Both vaccination regimens resulted in about 50% protection from malaria infection. And the researchers identified markers in each group that were associated with protection.
In the RRR group, circumsporozoite protein-specific antibody titers, prior to the challenge with infected mosquitoes, were associated with protection from malaria infection.
In addition, molecular signatures of B and plasma cells detected in peripheral blood mononuclear cells were associated with pre-challenge antibody titers and protection from malaria infection in the RRR group.
In the ARR group, protection from infection was associated with polyfunctional CD4+ T-cell responses 2 weeks after priming with Ad35, early signatures of innate immunity, and dendritic cell activation.
In both groups, natural killer (NK) cell signatures were negatively correlated with protection.
“Many of the genes contained in the predictive signatures are known to be expressed in natural killer cells, which mediate critical immune functions against viruses,” said study author Bali Pulendran, PhD, of Emory University School of Medicine in Atlanta, Georgia.
“It was a surprise to see such a robust ‘NK cell signature’ in predicting success of vaccination against the malaria parasite and raises the hypothesis that such cells may be playing a vital role in orchestrating immunity against malaria.”
Dr Pulendran and his colleagues said the results of this research suggest protective immunity against P falciparum can be achieved via multiple mechanisms.
“The extent to which these candidate signatures of protection can successfully predict vaccine efficacy in other field trials remain to be determined,” Dr Pulendran noted. ![]()
Making single-cell RNA sequencing widely available
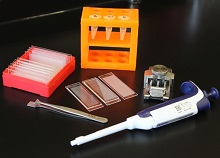
portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()

portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()

portable technology, Seq-Well, that
can prepare the RNA of many cells
for simultaneous sequencing.
Photo courtesy of Alex K. Shalek
and his colleagues
Researchers say they have developed a portable, low-cost platform for high-throughput, single-cell RNA sequencing.
The
team believes the technology, known as Seq-Well, could allow
scientists to more easily identify different cell types in blood and tissue
samples, helping them study how cancer cells respond to treatment, among other applications.
“Rather than trying to pick one marker that defines a cell type, using single-cell RNA sequencing, we can go in and look at everything a cell is expressing at a given moment,” said Alex K. Shalek, PhD, of the Massachusetts Institute of Technology in Cambridge.
“By finding common patterns across cells, we can figure out who those cells are.”
Dr Shalek and his colleagues have spent the past several years developing single-cell RNA sequencing strategies.
Now, they’ve created a new version of the technology that, they say, can rapidly analyze large numbers of cells using simple equipment.
“We’ve combined [Dr Shalek’s] technologies with some of ours in a way that makes it really accessible for researchers who want to do this type of sequencing on a range of different clinical samples and settings,” said J. Christopher Love, PhD, also of the Massachusetts Institute of Technology.
“It overcomes some of the barriers that are facing the adoption of these techniques more broadly.”
Drs Love and Shalek are the senior authors of a paper describing Seq-Well in Nature Methods.
Improving analysis
Key to sequencing RNA from large populations of cells is keeping track of which RNA transcripts came from which cell. The earliest techniques for this required sorting the cells into individual tubes or compartments of multiwell plates and then separately transforming each into a sequencing library.
That process works well but can’t handle large samples containing thousands of cells, such as blood samples or tissue biopsies, and costs between $25 and $35 per cell.
Dr Shalek and others have recently developed microfluidic techniques to help automate and parallelize the process considerably, but the amount of equipment required makes it impossible to be easily transported.
Drs Shalek and Love realized that technology Dr Love had previously developed to analyze protein secretions from single cells could be adapted to do single-cell RNA sequencing rapidly and inexpensively using a portable device.
Over the past several years, Dr Love’s lab has developed a microscale system that can isolate individual cells and measure the antibodies and other proteins that each cell secretes. The device resembles a tiny ice cube tray, with individual compartments for each cell.
Dr Love also developed a process known as microengraving that uses these trays, which can hold tens of thousands of cells, to measure each cell’s protein secretions.
To use this approach for sequencing RNA, the researchers created arrays of nanowells that each capture a single cell plus a barcoded bead to capture the RNA fragments.
The nanowells are sealed with a semipermeable membrane that allows the passage of chemicals needed to break the cells apart, while the RNA stays contained.
After the RNA binds to the beads, it is removed and sequenced. Using this process, the cost per cell is less than $1.
Uncovering unknowns
Similar to previous single-cell RNA sequencing techniques, the Seq-Well process captures and analyzes about 10% to 15% of the total number of RNA transcripts per cell.
“That is still a very rich set of information that maps to several thousand genes,” Dr Love said. “If you look at sets of these genes, you can start to understand the identity of those cells based on the sets of genes that are expressed in common.”
The researchers used Seq-Well to analyze macrophages infected with tuberculosis, allowing them to identify different pre-existing populations and responses to infection.
Dr Shalek and members of his lab also brought the technology to South Africa and analyzed tissue samples from tuberculosis- and HIV-infected patients there.
“Having a simple system that can go everywhere, I think, is going to be incredibly empowering,” Dr Shalek said.
Dr Love’s lab is now using this approach to analyze immune cells from people with food allergies, which could help researchers determine why some people are more likely to respond well to therapies designed to treat their allergies.
“There are still a lot of unknowns in chronic diseases, and these types of tools help you uncover new insights,” Dr Love said.
The research team has also joined forces with clinical investigators at Dana-Farber/Harvard Cancer Center to apply this technology toward the discovery of new combination immunotherapies for cancers. ![]()
Why like attracts like in malaria

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria. ![]()

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria. ![]()

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria.
Study shows no increased risk of mutations with iPSCs

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.”

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.”

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.”
Resistant malaria parasite spreading through Asia
Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.”

Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.”

Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.”
US-trained docs have higher patient death rate

Photo courtesy of the CDC
A large study has revealed a lower death rate among US patients treated by internationally trained doctors rather than US-trained doctors.
Researchers
analyzed data on more than 1.2 million US hospital admissions and found
a slight but statistically significant difference in 30-day mortality
for patients treated by internationally trained doctors and US-trained
doctors—11.2% and
11.6%, respectively (P<0.001).
These findings were published in The BMJ.
Yusuke Tsugawa, MD, PhD, of Harvard T H Chan School of Public Health in Boston, Massachusetts, and his colleagues conducted this research.
The team wanted to determine whether patient outcomes differ between general internists who graduated from a medical school outside the US and those who graduated from a US medical school.
The researchers analyzed data on the treatment of Medicare beneficiaries (age 65 and older) who were admitted to a hospital with a medical condition from 2011 through 2014. This included 1,215,490 hospital admissions and 44,227 general internists.
The primary outcome was 30-day patient mortality. Secondary outcomes were 30-day readmission rates and costs of care.
Compared with patients treated by US graduates, patients treated by international graduates had slightly more chronic conditions.
After adjusting for factors that could have affected the results (including patient characteristics, physician characteristics, and hospital fixed effects), the researchers found that patients cared for by international graduates had a lower rate of 30-day mortality than patients cared for by US graduates (11.2% and
11.6%, respectively, P<0.001).
The researchers said that for every 250 patients treated by US medical graduates, 1 patient’s life would be saved if the quality of care were equivalent between the international graduates and US graduates.
Thirty-day readmission rates did not differ significantly between the 2 types of graduates—15.4% for international graduates and 15.5% for US graduates (P=0.54).
However, the cost of care per admission was higher for international medical graduates—$1145 vs $1098 (P<0.001).
Further analysis to test the strength of these results made no difference to the overall findings.
One possible explanation for these findings, according to the researchers, is that the current approach for allowing international medical graduates to practice in the US may select for, on average, better physicians.
The team stressed that this is an observational study, so no firm conclusions can be drawn about cause and effect. Nevertheless, they said their findings “should reassure policymakers and the public that our current approach to licensing international medical graduates in the US is sufficiently rigorous to ensure high quality care.”

Photo courtesy of the CDC
A large study has revealed a lower death rate among US patients treated by internationally trained doctors rather than US-trained doctors.
Researchers
analyzed data on more than 1.2 million US hospital admissions and found
a slight but statistically significant difference in 30-day mortality
for patients treated by internationally trained doctors and US-trained
doctors—11.2% and
11.6%, respectively (P<0.001).
These findings were published in The BMJ.
Yusuke Tsugawa, MD, PhD, of Harvard T H Chan School of Public Health in Boston, Massachusetts, and his colleagues conducted this research.
The team wanted to determine whether patient outcomes differ between general internists who graduated from a medical school outside the US and those who graduated from a US medical school.
The researchers analyzed data on the treatment of Medicare beneficiaries (age 65 and older) who were admitted to a hospital with a medical condition from 2011 through 2014. This included 1,215,490 hospital admissions and 44,227 general internists.
The primary outcome was 30-day patient mortality. Secondary outcomes were 30-day readmission rates and costs of care.
Compared with patients treated by US graduates, patients treated by international graduates had slightly more chronic conditions.
After adjusting for factors that could have affected the results (including patient characteristics, physician characteristics, and hospital fixed effects), the researchers found that patients cared for by international graduates had a lower rate of 30-day mortality than patients cared for by US graduates (11.2% and
11.6%, respectively, P<0.001).
The researchers said that for every 250 patients treated by US medical graduates, 1 patient’s life would be saved if the quality of care were equivalent between the international graduates and US graduates.
Thirty-day readmission rates did not differ significantly between the 2 types of graduates—15.4% for international graduates and 15.5% for US graduates (P=0.54).
However, the cost of care per admission was higher for international medical graduates—$1145 vs $1098 (P<0.001).
Further analysis to test the strength of these results made no difference to the overall findings.
One possible explanation for these findings, according to the researchers, is that the current approach for allowing international medical graduates to practice in the US may select for, on average, better physicians.
The team stressed that this is an observational study, so no firm conclusions can be drawn about cause and effect. Nevertheless, they said their findings “should reassure policymakers and the public that our current approach to licensing international medical graduates in the US is sufficiently rigorous to ensure high quality care.”

Photo courtesy of the CDC
A large study has revealed a lower death rate among US patients treated by internationally trained doctors rather than US-trained doctors.
Researchers
analyzed data on more than 1.2 million US hospital admissions and found
a slight but statistically significant difference in 30-day mortality
for patients treated by internationally trained doctors and US-trained
doctors—11.2% and
11.6%, respectively (P<0.001).
These findings were published in The BMJ.
Yusuke Tsugawa, MD, PhD, of Harvard T H Chan School of Public Health in Boston, Massachusetts, and his colleagues conducted this research.
The team wanted to determine whether patient outcomes differ between general internists who graduated from a medical school outside the US and those who graduated from a US medical school.
The researchers analyzed data on the treatment of Medicare beneficiaries (age 65 and older) who were admitted to a hospital with a medical condition from 2011 through 2014. This included 1,215,490 hospital admissions and 44,227 general internists.
The primary outcome was 30-day patient mortality. Secondary outcomes were 30-day readmission rates and costs of care.
Compared with patients treated by US graduates, patients treated by international graduates had slightly more chronic conditions.
After adjusting for factors that could have affected the results (including patient characteristics, physician characteristics, and hospital fixed effects), the researchers found that patients cared for by international graduates had a lower rate of 30-day mortality than patients cared for by US graduates (11.2% and
11.6%, respectively, P<0.001).
The researchers said that for every 250 patients treated by US medical graduates, 1 patient’s life would be saved if the quality of care were equivalent between the international graduates and US graduates.
Thirty-day readmission rates did not differ significantly between the 2 types of graduates—15.4% for international graduates and 15.5% for US graduates (P=0.54).
However, the cost of care per admission was higher for international medical graduates—$1145 vs $1098 (P<0.001).
Further analysis to test the strength of these results made no difference to the overall findings.
One possible explanation for these findings, according to the researchers, is that the current approach for allowing international medical graduates to practice in the US may select for, on average, better physicians.
The team stressed that this is an observational study, so no firm conclusions can be drawn about cause and effect. Nevertheless, they said their findings “should reassure policymakers and the public that our current approach to licensing international medical graduates in the US is sufficiently rigorous to ensure high quality care.”





